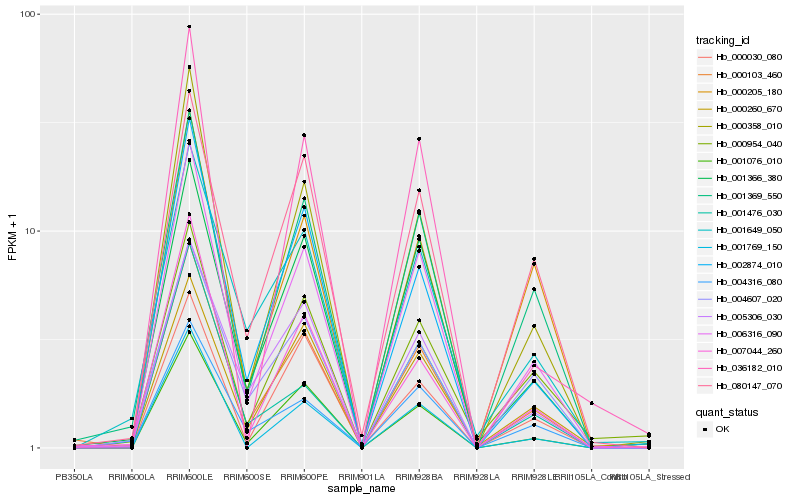
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006316_090 |
0.0 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 2 |
Hb_000358_010 |
0.0642152879 |
- |
- |
PREDICTED: basic blue protein [Populus euphratica] |
| 3 |
Hb_000103_460 |
0.0671767056 |
- |
- |
PREDICTED: calmodulin-like protein 11 [Jatropha curcas] |
| 4 |
Hb_001076_010 |
0.0769088479 |
transcription factor |
TF Family: B3 |
DNA binding protein, putative [Ricinus communis] |
| 5 |
Hb_001369_550 |
0.0847807861 |
- |
- |
21 kDa protein precursor, putative [Ricinus communis] |
| 6 |
Hb_001366_380 |
0.0897472248 |
- |
- |
PREDICTED: pectin acetylesterase 6-like isoform X2 [Jatropha curcas] |
| 7 |
Hb_004316_080 |
0.095717033 |
- |
- |
PREDICTED: uncharacterized protein LOC104602212 [Nelumbo nucifera] |
| 8 |
Hb_002874_010 |
0.0989605601 |
- |
- |
PREDICTED: replication protein A 32 kDa subunit A isoform X1 [Jatropha curcas] |
| 9 |
Hb_001769_150 |
0.101992305 |
- |
- |
PREDICTED: uncharacterized protein LOC105633924 [Jatropha curcas] |
| 10 |
Hb_007044_260 |
0.1075627842 |
- |
- |
PREDICTED: uncharacterized protein LOC105638706 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000205_180 |
0.1133578744 |
- |
- |
lipoxygenase, putative [Ricinus communis] |
| 12 |
Hb_080147_070 |
0.1167920375 |
- |
- |
Rop5 [Hevea brasiliensis] |
| 13 |
Hb_000260_670 |
0.1171806653 |
- |
- |
PREDICTED: serine/threonine-protein kinase TIO isoform X1 [Jatropha curcas] |
| 14 |
Hb_005306_030 |
0.1176473156 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1 [Jatropha curcas] |
| 15 |
Hb_036182_010 |
0.1248298635 |
- |
- |
hypothetical protein POPTR_0473s00220g [Populus trichocarpa] |
| 16 |
Hb_000954_040 |
0.125371741 |
- |
- |
PREDICTED: metal tolerance protein 4 [Jatropha curcas] |
| 17 |
Hb_001649_050 |
0.1273091825 |
- |
- |
Shaggy-like kinase 13 isoform 1 [Theobroma cacao] |
| 18 |
Hb_001476_030 |
0.1275864813 |
- |
- |
Cationic peroxidase 2 precursor, putative [Ricinus communis] |
| 19 |
Hb_000030_080 |
0.1283386784 |
- |
- |
PREDICTED: uncharacterized protein LOC105632640 [Jatropha curcas] |
| 20 |
Hb_004607_020 |
0.1287428498 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER-like [Jatropha curcas] |