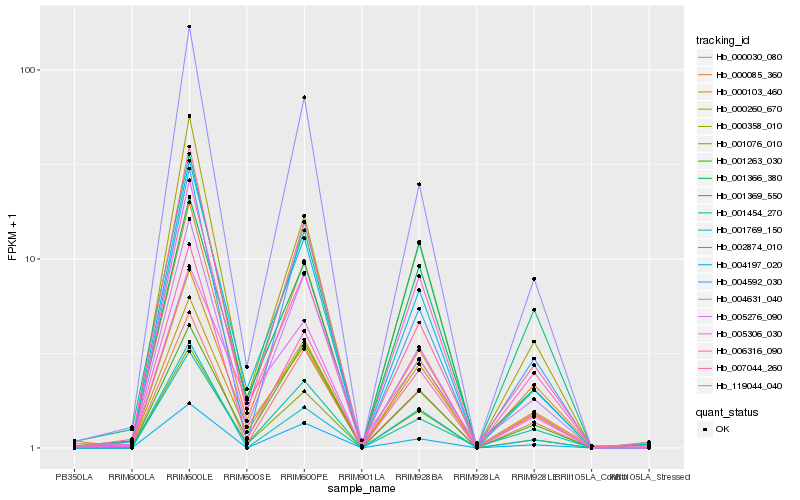
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001076_010 |
0.0 |
transcription factor |
TF Family: B3 |
DNA binding protein, putative [Ricinus communis] |
| 2 |
Hb_002874_010 |
0.0604281196 |
- |
- |
PREDICTED: replication protein A 32 kDa subunit A isoform X1 [Jatropha curcas] |
| 3 |
Hb_000358_010 |
0.0677800428 |
- |
- |
PREDICTED: basic blue protein [Populus euphratica] |
| 4 |
Hb_006316_090 |
0.0769088479 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 5 |
Hb_004631_040 |
0.0783947178 |
- |
- |
hypothetical protein CARUB_v10021660mg, partial [Capsella rubella] |
| 6 |
Hb_000030_080 |
0.0804120827 |
- |
- |
PREDICTED: uncharacterized protein LOC105632640 [Jatropha curcas] |
| 7 |
Hb_000103_460 |
0.0887396314 |
- |
- |
PREDICTED: calmodulin-like protein 11 [Jatropha curcas] |
| 8 |
Hb_007044_260 |
0.0975379649 |
- |
- |
PREDICTED: uncharacterized protein LOC105638706 isoform X1 [Jatropha curcas] |
| 9 |
Hb_004592_030 |
0.1016919607 |
- |
- |
unknown [Lotus japonicus] |
| 10 |
Hb_004197_020 |
0.1023064946 |
- |
- |
Anthocyanin 5-aromatic acyltransferase, putative [Ricinus communis] |
| 11 |
Hb_001366_380 |
0.1047372176 |
- |
- |
PREDICTED: pectin acetylesterase 6-like isoform X2 [Jatropha curcas] |
| 12 |
Hb_000085_360 |
0.1077166704 |
- |
- |
PREDICTED: kinesin-like protein KIN12B [Jatropha curcas] |
| 13 |
Hb_005306_030 |
0.1110292011 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1 [Jatropha curcas] |
| 14 |
Hb_001369_550 |
0.1135908152 |
- |
- |
21 kDa protein precursor, putative [Ricinus communis] |
| 15 |
Hb_000260_670 |
0.1140724736 |
- |
- |
PREDICTED: serine/threonine-protein kinase TIO isoform X1 [Jatropha curcas] |
| 16 |
Hb_001769_150 |
0.1161437065 |
- |
- |
PREDICTED: uncharacterized protein LOC105633924 [Jatropha curcas] |
| 17 |
Hb_001454_270 |
0.1164302532 |
- |
- |
PREDICTED: condensin-2 complex subunit D3 isoform X2 [Jatropha curcas] |
| 18 |
Hb_119044_040 |
0.1201279392 |
- |
- |
cyclin B, putative [Ricinus communis] |
| 19 |
Hb_005276_090 |
0.1211639099 |
- |
- |
PREDICTED: uncharacterized protein LOC105649997 isoform X2 [Jatropha curcas] |
| 20 |
Hb_001263_030 |
0.1213263916 |
- |
- |
PREDICTED: uncharacterized protein At3g28850-like [Jatropha curcas] |