| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
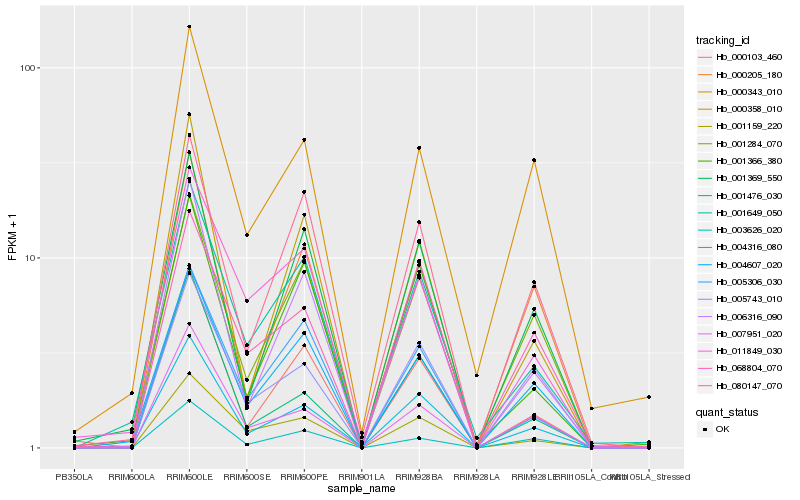
Hb_004316_080 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104602212 [Nelumbo nucifera] |
| 2 |
Hb_006316_090 |
0.095717033 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 3 |
Hb_004607_020 |
0.1022367589 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER-like [Jatropha curcas] |
| 4 |
Hb_001649_050 |
0.1083375988 |
- |
- |
Shaggy-like kinase 13 isoform 1 [Theobroma cacao] |
| 5 |
Hb_007951_020 |
0.1088981135 |
- |
- |
cationic amino acid transporter 5 family protein [Populus trichocarpa] |
| 6 |
Hb_005743_010 |
0.1094626574 |
- |
- |
RING/U-box superfamily protein, putative [Theobroma cacao] |
| 7 |
Hb_001476_030 |
0.1107125166 |
- |
- |
Cationic peroxidase 2 precursor, putative [Ricinus communis] |
| 8 |
Hb_001369_550 |
0.1117837416 |
- |
- |
21 kDa protein precursor, putative [Ricinus communis] |
| 9 |
Hb_000103_460 |
0.1127917178 |
- |
- |
PREDICTED: calmodulin-like protein 11 [Jatropha curcas] |
| 10 |
Hb_068804_070 |
0.1198902052 |
- |
- |
hypothetical protein JCGZ_12627 [Jatropha curcas] |
| 11 |
Hb_001284_070 |
0.1202888589 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000205_180 |
0.1236034499 |
- |
- |
lipoxygenase, putative [Ricinus communis] |
| 13 |
Hb_001366_380 |
0.1250036447 |
- |
- |
PREDICTED: pectin acetylesterase 6-like isoform X2 [Jatropha curcas] |
| 14 |
Hb_080147_070 |
0.1265080264 |
- |
- |
Rop5 [Hevea brasiliensis] |
| 15 |
Hb_000343_010 |
0.1272089745 |
- |
- |
F3F9.11 [Arabidopsis thaliana] |
| 16 |
Hb_001159_220 |
0.1301050562 |
- |
- |
ESC, putative [Ricinus communis] |
| 17 |
Hb_005306_030 |
0.1306480993 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1 [Jatropha curcas] |
| 18 |
Hb_003626_020 |
0.1351357887 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At5g24080 [Jatropha curcas] |
| 19 |
Hb_011849_030 |
0.1361652051 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000358_010 |
0.1377828444 |
- |
- |
PREDICTED: basic blue protein [Populus euphratica] |