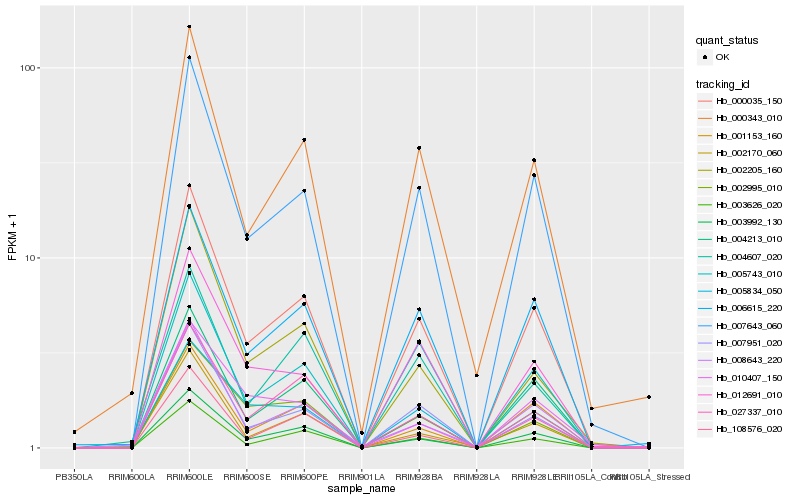
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000035_150 |
0.0 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 40 [Jatropha curcas] |
| 2 |
Hb_007643_060 |
0.046761975 |
- |
- |
- |
| 3 |
Hb_003992_130 |
0.0632485114 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 4 |
Hb_007951_020 |
0.0750004923 |
- |
- |
cationic amino acid transporter 5 family protein [Populus trichocarpa] |
| 5 |
Hb_006615_220 |
0.0869035804 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor BIM1 isoform X2 [Jatropha curcas] |
| 6 |
Hb_003626_020 |
0.0893599875 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At5g24080 [Jatropha curcas] |
| 7 |
Hb_004213_010 |
0.0924729601 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 8 |
Hb_001153_160 |
0.0942456106 |
- |
- |
molybdopterin cofactor sulfurase, putative [Ricinus communis] |
| 9 |
Hb_000343_010 |
0.0990885115 |
- |
- |
F3F9.11 [Arabidopsis thaliana] |
| 10 |
Hb_002205_160 |
0.1062055824 |
- |
- |
chalcone isomerase [Hibiscus cannabinus] |
| 11 |
Hb_012691_010 |
0.1084160318 |
- |
- |
hypothetical protein POPTR_0005s18750g [Populus trichocarpa] |
| 12 |
Hb_005834_050 |
0.1118420255 |
- |
- |
PREDICTED: cationic amino acid transporter 1-like [Jatropha curcas] |
| 13 |
Hb_005743_010 |
0.1123096176 |
- |
- |
RING/U-box superfamily protein, putative [Theobroma cacao] |
| 14 |
Hb_002995_010 |
0.1127249968 |
- |
- |
PREDICTED: flavonol synthase/flavanone 3-hydroxylase [Jatropha curcas] |
| 15 |
Hb_008643_220 |
0.1182939924 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g53440 [Jatropha curcas] |
| 16 |
Hb_027337_010 |
0.1204827981 |
- |
- |
Potassium transporter, putative [Ricinus communis] |
| 17 |
Hb_004607_020 |
0.1235005438 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER-like [Jatropha curcas] |
| 18 |
Hb_108576_020 |
0.1236150664 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002170_060 |
0.1247514645 |
- |
- |
PREDICTED: mitogen-activated protein kinase-binding protein 1 [Jatropha curcas] |
| 20 |
Hb_010407_150 |
0.1292734906 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0008s16660g [Populus trichocarpa] |