| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
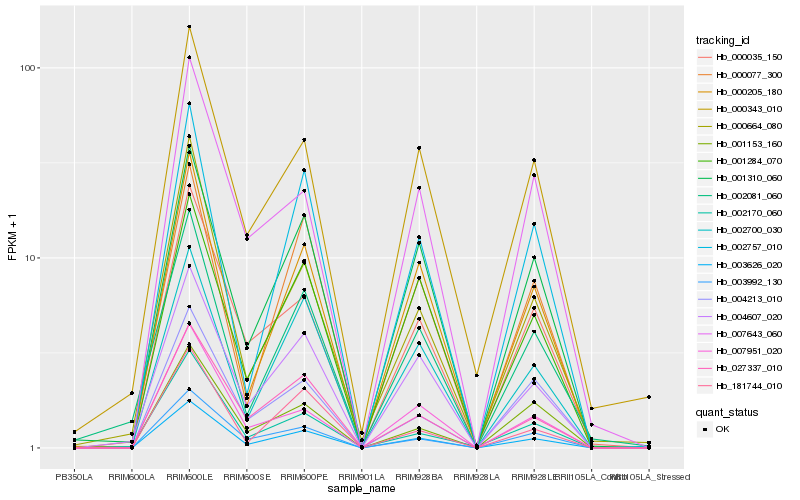
Hb_003626_020 |
0.0 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At5g24080 [Jatropha curcas] |
| 2 |
Hb_000205_180 |
0.0805341313 |
- |
- |
lipoxygenase, putative [Ricinus communis] |
| 3 |
Hb_003992_130 |
0.0818176602 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 4 |
Hb_004607_020 |
0.0873387289 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER-like [Jatropha curcas] |
| 5 |
Hb_000035_150 |
0.0893599875 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 40 [Jatropha curcas] |
| 6 |
Hb_027337_010 |
0.0944121053 |
- |
- |
Potassium transporter, putative [Ricinus communis] |
| 7 |
Hb_001310_060 |
0.0990632455 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 8 |
Hb_007951_020 |
0.0990907835 |
- |
- |
cationic amino acid transporter 5 family protein [Populus trichocarpa] |
| 9 |
Hb_002700_030 |
0.1005600035 |
- |
- |
PREDICTED: serine carboxypeptidase-like 45 [Jatropha curcas] |
| 10 |
Hb_002757_010 |
0.103716368 |
- |
- |
hypothetical protein POPTR_0013s10580g [Populus trichocarpa] |
| 11 |
Hb_001284_070 |
0.1044530545 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000664_080 |
0.1063544564 |
- |
- |
PREDICTED: NADP-dependent malic enzyme isoform X2 [Jatropha curcas] |
| 13 |
Hb_000343_010 |
0.1094703039 |
- |
- |
F3F9.11 [Arabidopsis thaliana] |
| 14 |
Hb_004213_010 |
0.1127963645 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 15 |
Hb_001153_160 |
0.1130752571 |
- |
- |
molybdopterin cofactor sulfurase, putative [Ricinus communis] |
| 16 |
Hb_002170_060 |
0.1137610367 |
- |
- |
PREDICTED: mitogen-activated protein kinase-binding protein 1 [Jatropha curcas] |
| 17 |
Hb_181744_010 |
0.1139434199 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 18 |
Hb_002081_060 |
0.1157494683 |
- |
- |
cyclin d, putative [Ricinus communis] |
| 19 |
Hb_007643_060 |
0.1169098758 |
- |
- |
- |
| 20 |
Hb_000077_300 |
0.1179745723 |
- |
- |
PREDICTED: expansin-B3-like [Jatropha curcas] |