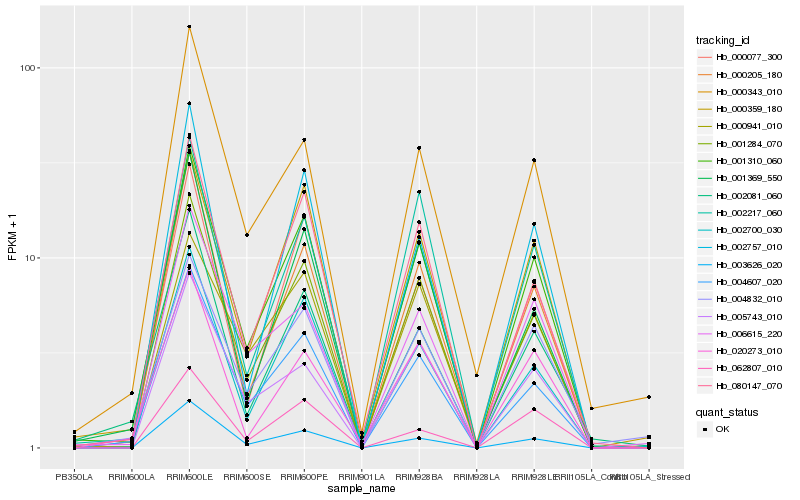
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001310_060 |
0.0 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 2 |
Hb_001284_070 |
0.0550985753 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_080147_070 |
0.0795965503 |
- |
- |
Rop5 [Hevea brasiliensis] |
| 4 |
Hb_004607_020 |
0.0806442505 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER-like [Jatropha curcas] |
| 5 |
Hb_000205_180 |
0.0814221919 |
- |
- |
lipoxygenase, putative [Ricinus communis] |
| 6 |
Hb_002700_030 |
0.0827052628 |
- |
- |
PREDICTED: serine carboxypeptidase-like 45 [Jatropha curcas] |
| 7 |
Hb_000077_300 |
0.0907524217 |
- |
- |
PREDICTED: expansin-B3-like [Jatropha curcas] |
| 8 |
Hb_000359_180 |
0.0914826473 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_003626_020 |
0.0990632455 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At5g24080 [Jatropha curcas] |
| 10 |
Hb_004832_010 |
0.0991122082 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001369_550 |
0.1046183189 |
- |
- |
21 kDa protein precursor, putative [Ricinus communis] |
| 12 |
Hb_002757_010 |
0.1047325445 |
- |
- |
hypothetical protein POPTR_0013s10580g [Populus trichocarpa] |
| 13 |
Hb_006615_220 |
0.1068036418 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor BIM1 isoform X2 [Jatropha curcas] |
| 14 |
Hb_002081_060 |
0.1082020208 |
- |
- |
cyclin d, putative [Ricinus communis] |
| 15 |
Hb_002217_060 |
0.1090122219 |
- |
- |
PREDICTED: polygalacturonase At1g48100 [Jatropha curcas] |
| 16 |
Hb_000343_010 |
0.1111175153 |
- |
- |
F3F9.11 [Arabidopsis thaliana] |
| 17 |
Hb_005743_010 |
0.1115959692 |
- |
- |
RING/U-box superfamily protein, putative [Theobroma cacao] |
| 18 |
Hb_000941_010 |
0.1182718875 |
- |
- |
hypothetical protein RCOM_0504460 [Ricinus communis] |
| 19 |
Hb_062807_010 |
0.1210138816 |
- |
- |
hypothetical protein JCGZ_15896 [Jatropha curcas] |
| 20 |
Hb_020273_010 |
0.1231631828 |
- |
- |
PREDICTED: beta-galactosidase 3 [Jatropha curcas] |