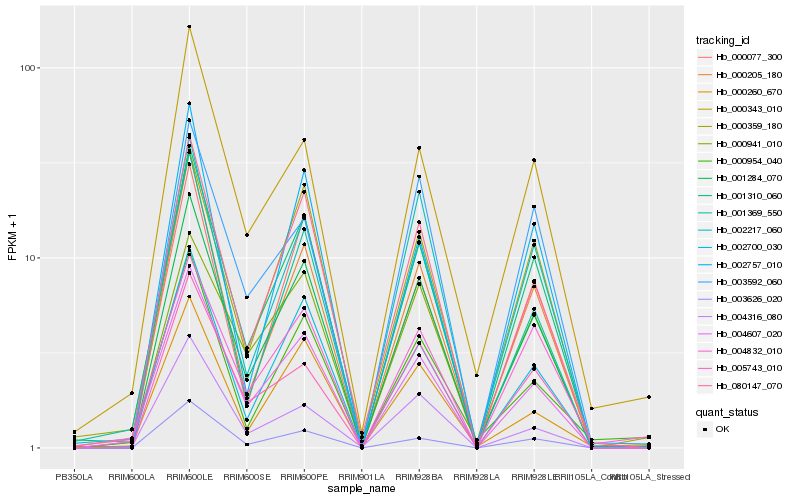
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001284_070 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001310_060 |
0.0550985753 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 3 |
Hb_080147_070 |
0.0650785725 |
- |
- |
Rop5 [Hevea brasiliensis] |
| 4 |
Hb_004607_020 |
0.0782373976 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER-like [Jatropha curcas] |
| 5 |
Hb_002700_030 |
0.07960312 |
- |
- |
PREDICTED: serine carboxypeptidase-like 45 [Jatropha curcas] |
| 6 |
Hb_000205_180 |
0.0819225697 |
- |
- |
lipoxygenase, putative [Ricinus communis] |
| 7 |
Hb_001369_550 |
0.0947922763 |
- |
- |
21 kDa protein precursor, putative [Ricinus communis] |
| 8 |
Hb_002217_060 |
0.0997647302 |
- |
- |
PREDICTED: polygalacturonase At1g48100 [Jatropha curcas] |
| 9 |
Hb_000077_300 |
0.1031759471 |
- |
- |
PREDICTED: expansin-B3-like [Jatropha curcas] |
| 10 |
Hb_005743_010 |
0.1039811963 |
- |
- |
RING/U-box superfamily protein, putative [Theobroma cacao] |
| 11 |
Hb_003626_020 |
0.1044530545 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At5g24080 [Jatropha curcas] |
| 12 |
Hb_000359_180 |
0.1083312004 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000260_670 |
0.1100380636 |
- |
- |
PREDICTED: serine/threonine-protein kinase TIO isoform X1 [Jatropha curcas] |
| 14 |
Hb_000941_010 |
0.1134820703 |
- |
- |
hypothetical protein RCOM_0504460 [Ricinus communis] |
| 15 |
Hb_002757_010 |
0.1175406139 |
- |
- |
hypothetical protein POPTR_0013s10580g [Populus trichocarpa] |
| 16 |
Hb_004316_080 |
0.1202888589 |
- |
- |
PREDICTED: uncharacterized protein LOC104602212 [Nelumbo nucifera] |
| 17 |
Hb_000954_040 |
0.1211847532 |
- |
- |
PREDICTED: metal tolerance protein 4 [Jatropha curcas] |
| 18 |
Hb_000343_010 |
0.1216641773 |
- |
- |
F3F9.11 [Arabidopsis thaliana] |
| 19 |
Hb_004832_010 |
0.1275383342 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_003592_060 |
0.1293919759 |
- |
- |
hypothetical protein F383_11017 [Gossypium arboreum] |