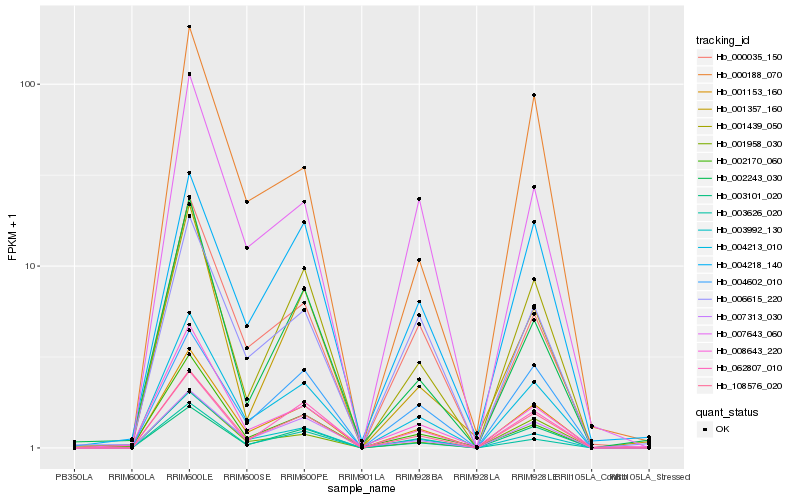
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001153_160 |
0.0 |
- |
- |
molybdopterin cofactor sulfurase, putative [Ricinus communis] |
| 2 |
Hb_004213_010 |
0.0052455288 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 3 |
Hb_108576_020 |
0.0329303498 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_003992_130 |
0.0678654027 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 5 |
Hb_000035_150 |
0.0942456106 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 40 [Jatropha curcas] |
| 6 |
Hb_007313_030 |
0.0953608757 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK2 [Jatropha curcas] |
| 7 |
Hb_003101_020 |
0.0958304164 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor NAM-B2 [Jatropha curcas] |
| 8 |
Hb_001439_050 |
0.0970725023 |
- |
- |
plastocyanin-like domain-containing family protein [Populus trichocarpa] |
| 9 |
Hb_062807_010 |
0.1025699944 |
- |
- |
hypothetical protein JCGZ_15896 [Jatropha curcas] |
| 10 |
Hb_001958_030 |
0.1064755233 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4-like [Jatropha curcas] |
| 11 |
Hb_007643_060 |
0.1085434051 |
- |
- |
- |
| 12 |
Hb_003626_020 |
0.1130752571 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At5g24080 [Jatropha curcas] |
| 13 |
Hb_006615_220 |
0.1145103434 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor BIM1 isoform X2 [Jatropha curcas] |
| 14 |
Hb_004602_010 |
0.123323079 |
- |
- |
PREDICTED: uncharacterized protein LOC105042236 isoform X2 [Elaeis guineensis] |
| 15 |
Hb_002170_060 |
0.1239985499 |
- |
- |
PREDICTED: mitogen-activated protein kinase-binding protein 1 [Jatropha curcas] |
| 16 |
Hb_008643_220 |
0.1240534762 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g53440 [Jatropha curcas] |
| 17 |
Hb_002243_030 |
0.1278566151 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 3-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_000188_070 |
0.1309331174 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 19 |
Hb_001357_160 |
0.1349265967 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_004218_140 |
0.135376643 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD11 isoform X3 [Jatropha curcas] |