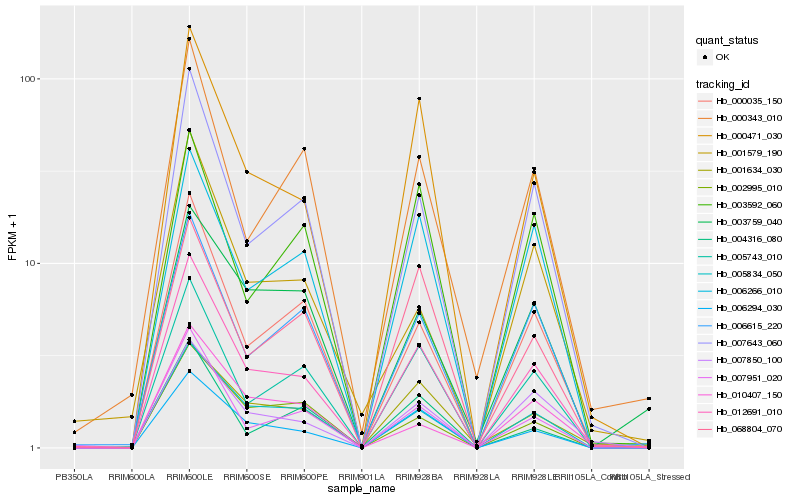
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012691_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0005s18750g [Populus trichocarpa] |
| 2 |
Hb_006294_030 |
0.0882590669 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 3 |
Hb_000471_030 |
0.0888407533 |
- |
- |
- |
| 4 |
Hb_007643_060 |
0.0903721128 |
- |
- |
- |
| 5 |
Hb_005834_050 |
0.09949141 |
- |
- |
PREDICTED: cationic amino acid transporter 1-like [Jatropha curcas] |
| 6 |
Hb_007951_020 |
0.1006130218 |
- |
- |
cationic amino acid transporter 5 family protein [Populus trichocarpa] |
| 7 |
Hb_005743_010 |
0.1083442779 |
- |
- |
RING/U-box superfamily protein, putative [Theobroma cacao] |
| 8 |
Hb_000035_150 |
0.1084160318 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 40 [Jatropha curcas] |
| 9 |
Hb_006615_220 |
0.1179924963 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor BIM1 isoform X2 [Jatropha curcas] |
| 10 |
Hb_007850_100 |
0.1314412011 |
- |
- |
PREDICTED: uncharacterized protein LOC105640661 [Jatropha curcas] |
| 11 |
Hb_006266_010 |
0.1338172688 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 12 |
Hb_002995_010 |
0.134381442 |
- |
- |
PREDICTED: flavonol synthase/flavanone 3-hydroxylase [Jatropha curcas] |
| 13 |
Hb_000343_010 |
0.1377621731 |
- |
- |
F3F9.11 [Arabidopsis thaliana] |
| 14 |
Hb_068804_070 |
0.1404591827 |
- |
- |
hypothetical protein JCGZ_12627 [Jatropha curcas] |
| 15 |
Hb_001634_030 |
0.142086249 |
- |
- |
Receptor like protein 1, putative [Theobroma cacao] |
| 16 |
Hb_010407_150 |
0.1568793272 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0008s16660g [Populus trichocarpa] |
| 17 |
Hb_004316_080 |
0.1575624493 |
- |
- |
PREDICTED: uncharacterized protein LOC104602212 [Nelumbo nucifera] |
| 18 |
Hb_003592_060 |
0.159121271 |
- |
- |
hypothetical protein F383_11017 [Gossypium arboreum] |
| 19 |
Hb_001579_190 |
0.1593135726 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_003759_040 |
0.1596187311 |
- |
- |
PREDICTED: aldo-keto reductase family 4 member C9-like [Jatropha curcas] |