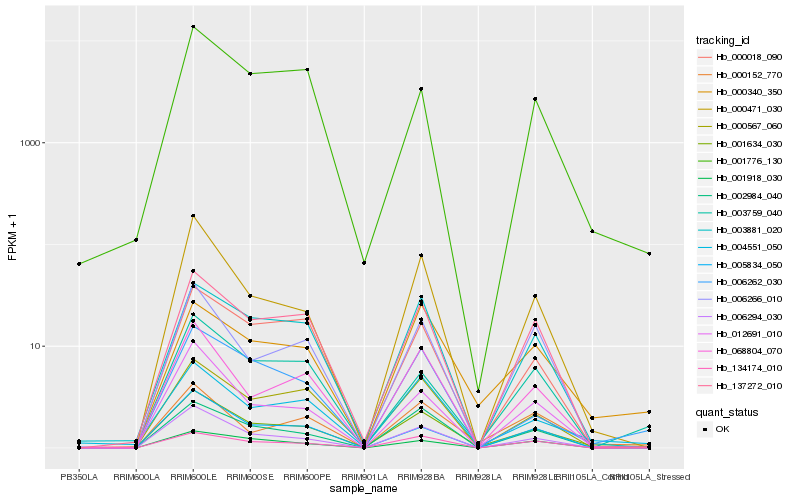
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001634_030 |
0.0 |
- |
- |
Receptor like protein 1, putative [Theobroma cacao] |
| 2 |
Hb_006294_030 |
0.1198372955 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 3 |
Hb_004551_050 |
0.1253706282 |
- |
- |
PREDICTED: glucose-6-phosphate/phosphate translocator 2, chloroplastic [Jatropha curcas] |
| 4 |
Hb_137272_010 |
0.128009887 |
- |
- |
PREDICTED: cysteine-rich receptor-like protein kinase 10 [Jatropha curcas] |
| 5 |
Hb_000567_060 |
0.128562644 |
- |
- |
PREDICTED: U-box domain-containing protein 7 [Jatropha curcas] |
| 6 |
Hb_068804_070 |
0.1333095727 |
- |
- |
hypothetical protein JCGZ_12627 [Jatropha curcas] |
| 7 |
Hb_003881_020 |
0.1349846753 |
- |
- |
PREDICTED: stellacyanin-like [Jatropha curcas] |
| 8 |
Hb_000471_030 |
0.1361785464 |
- |
- |
- |
| 9 |
Hb_134174_010 |
0.1405640107 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 10 |
Hb_012691_010 |
0.142086249 |
- |
- |
hypothetical protein POPTR_0005s18750g [Populus trichocarpa] |
| 11 |
Hb_002984_040 |
0.1421344046 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 12 |
Hb_000340_350 |
0.1450770405 |
- |
- |
PREDICTED: sugar transporter ERD6-like 7 isoform X1 [Jatropha curcas] |
| 13 |
Hb_006262_030 |
0.1467426611 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 14 |
Hb_000018_090 |
0.1485371845 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 15 |
Hb_000152_770 |
0.1523616643 |
- |
- |
PREDICTED: dynein light chain, cytoplasmic-like [Jatropha curcas] |
| 16 |
Hb_003759_040 |
0.1543699535 |
- |
- |
PREDICTED: aldo-keto reductase family 4 member C9-like [Jatropha curcas] |
| 17 |
Hb_005834_050 |
0.1585102559 |
- |
- |
PREDICTED: cationic amino acid transporter 1-like [Jatropha curcas] |
| 18 |
Hb_001776_130 |
0.1599093713 |
- |
- |
metallothionein [Hevea brasiliensis] |
| 19 |
Hb_006266_010 |
0.1619377972 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 20 |
Hb_001918_030 |
0.1623533838 |
- |
- |
- |