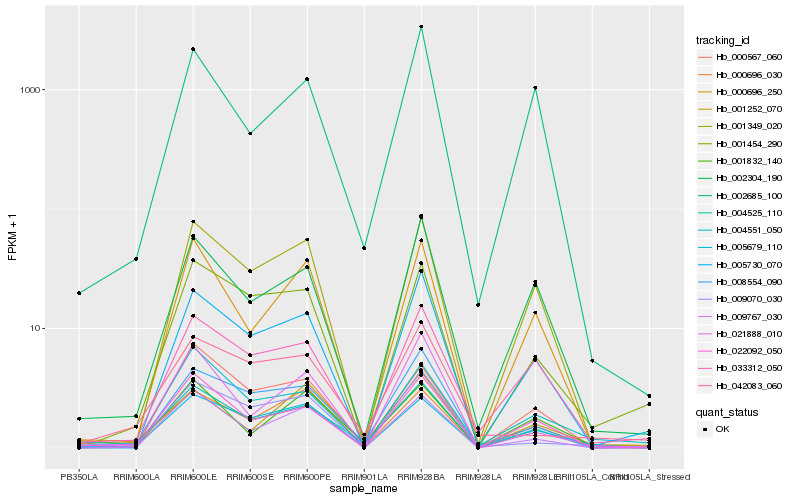
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004525_110 |
0.0 |
- |
- |
hypothetical protein POPTR_0007s12850g [Populus trichocarpa] |
| 2 |
Hb_008554_090 |
0.1258139744 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 3 |
Hb_022092_050 |
0.1296391314 |
- |
- |
PREDICTED: uncharacterized protein LOC105136214 [Populus euphratica] |
| 4 |
Hb_000696_030 |
0.1356097704 |
- |
- |
PREDICTED: uncharacterized protein LOC105647085 isoform X1 [Jatropha curcas] |
| 5 |
Hb_004551_050 |
0.1367522431 |
- |
- |
PREDICTED: glucose-6-phosphate/phosphate translocator 2, chloroplastic [Jatropha curcas] |
| 6 |
Hb_033312_050 |
0.1373894266 |
- |
- |
PREDICTED: uncharacterized protein LOC105638598, partial [Jatropha curcas] |
| 7 |
Hb_001349_020 |
0.1388338115 |
- |
- |
PREDICTED: leucoanthocyanidin reductase-like [Jatropha curcas] |
| 8 |
Hb_005679_110 |
0.1390093897 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0271670 [Jatropha curcas] |
| 9 |
Hb_002304_190 |
0.1409377879 |
- |
- |
PREDICTED: 3'-N-debenzoyl-2'-deoxytaxol N-benzoyltransferase [Jatropha curcas] |
| 10 |
Hb_000696_250 |
0.1472968764 |
- |
- |
PREDICTED: patellin-6 [Jatropha curcas] |
| 11 |
Hb_001454_290 |
0.1483977147 |
- |
- |
PREDICTED: uncharacterized protein LOC105643442 [Jatropha curcas] |
| 12 |
Hb_005730_070 |
0.1493247536 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: ABC transporter C family member 8 [Jatropha curcas] |
| 13 |
Hb_009070_030 |
0.1538836251 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 10 [Jatropha curcas] |
| 14 |
Hb_009767_030 |
0.1562057456 |
- |
- |
PREDICTED: uncharacterized protein LOC105631977 [Jatropha curcas] |
| 15 |
Hb_021888_010 |
0.1568685372 |
- |
- |
PREDICTED: aspartic proteinase-like protein 1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001252_070 |
0.1574837153 |
- |
- |
hypothetical protein JCGZ_01385 [Jatropha curcas] |
| 17 |
Hb_001832_140 |
0.1583225933 |
- |
- |
endomembrane protein 70 family protein [Medicago truncatula] |
| 18 |
Hb_002685_100 |
0.1591167632 |
- |
- |
low temperature inducible SLTI66 [Glycine max] |
| 19 |
Hb_042083_060 |
0.1602041378 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g45780 [Jatropha curcas] |
| 20 |
Hb_000567_060 |
0.1602186161 |
- |
- |
PREDICTED: U-box domain-containing protein 7 [Jatropha curcas] |