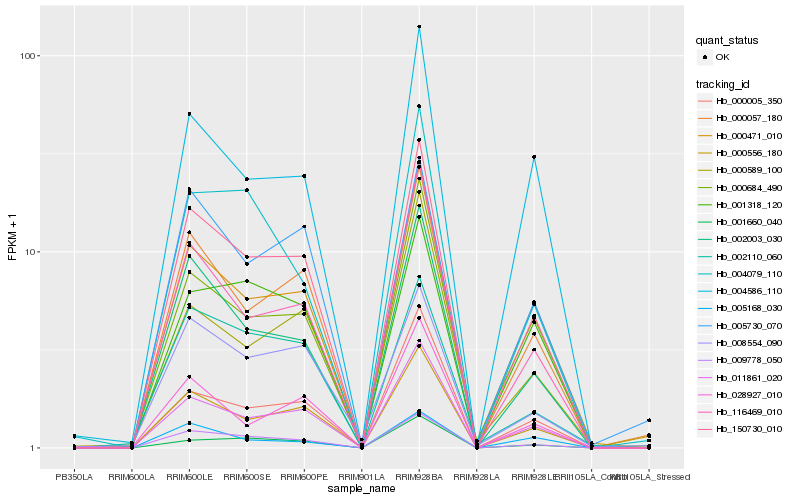
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_150730_010 |
0.0 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Jatropha curcas] |
| 2 |
Hb_011861_020 |
0.0610293052 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_06096 [Jatropha curcas] |
| 3 |
Hb_000057_180 |
0.0719505759 |
- |
- |
hypothetical protein POPTR_0001s18200g [Populus trichocarpa] |
| 4 |
Hb_002003_030 |
0.07556047 |
- |
- |
PREDICTED: UDP-glycosyltransferase 74F1-like [Jatropha curcas] |
| 5 |
Hb_009778_050 |
0.0770289921 |
- |
- |
hypothetical protein JCGZ_08917 [Jatropha curcas] |
| 6 |
Hb_116469_010 |
0.0792743202 |
- |
- |
PREDICTED: uncharacterized protein LOC105647834 [Jatropha curcas] |
| 7 |
Hb_000471_010 |
0.0845937143 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 8 |
Hb_000684_490 |
0.1046583871 |
transcription factor |
TF Family: WRKY |
PREDICTED: WRKY transcription factor 44 [Jatropha curcas] |
| 9 |
Hb_004586_110 |
0.1137742291 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 10 |
Hb_000005_350 |
0.1139245251 |
- |
- |
PREDICTED: uncharacterized protein LOC105634263 [Jatropha curcas] |
| 11 |
Hb_005730_070 |
0.1151835053 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: ABC transporter C family member 8 [Jatropha curcas] |
| 12 |
Hb_028927_010 |
0.1182229772 |
- |
- |
leucine-rich repeat protein, putative [Ricinus communis] |
| 13 |
Hb_001660_040 |
0.1222759334 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Jatropha curcas] |
| 14 |
Hb_008554_090 |
0.1271181037 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 15 |
Hb_002110_060 |
0.1288299861 |
- |
- |
PREDICTED: serine/threonine-protein kinase-like protein At1g28390 [Jatropha curcas] |
| 16 |
Hb_004079_110 |
0.1303418025 |
- |
- |
PREDICTED: uncharacterized protein LOC100257802 [Vitis vinifera] |
| 17 |
Hb_001318_120 |
0.1320097168 |
- |
- |
PREDICTED: uncharacterized protein LOC105642675 [Jatropha curcas] |
| 18 |
Hb_000589_100 |
0.1326728102 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 19 |
Hb_000556_180 |
0.1350206952 |
- |
- |
hypothetical protein POPTR_0014s06690g [Populus trichocarpa] |
| 20 |
Hb_005168_030 |
0.1419829548 |
- |
- |
unknown [Zea mays] |