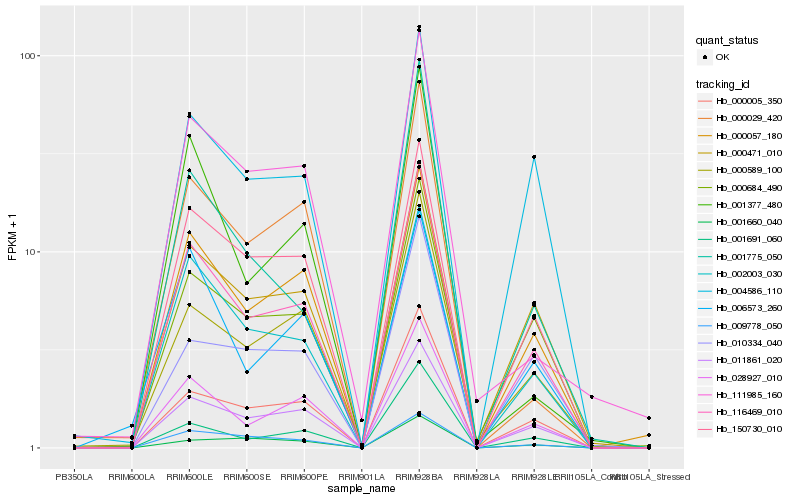
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_116469_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647834 [Jatropha curcas] |
| 2 |
Hb_011861_020 |
0.0662620923 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_06096 [Jatropha curcas] |
| 3 |
Hb_002003_030 |
0.0726719284 |
- |
- |
PREDICTED: UDP-glycosyltransferase 74F1-like [Jatropha curcas] |
| 4 |
Hb_028927_010 |
0.0772099401 |
- |
- |
leucine-rich repeat protein, putative [Ricinus communis] |
| 5 |
Hb_150730_010 |
0.0792743202 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Jatropha curcas] |
| 6 |
Hb_000057_180 |
0.0814798429 |
- |
- |
hypothetical protein POPTR_0001s18200g [Populus trichocarpa] |
| 7 |
Hb_000005_350 |
0.0854827846 |
- |
- |
PREDICTED: uncharacterized protein LOC105634263 [Jatropha curcas] |
| 8 |
Hb_000471_010 |
0.0877704929 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 9 |
Hb_000684_490 |
0.0965735578 |
transcription factor |
TF Family: WRKY |
PREDICTED: WRKY transcription factor 44 [Jatropha curcas] |
| 10 |
Hb_000589_100 |
0.1058751044 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 11 |
Hb_004586_110 |
0.1109363035 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 12 |
Hb_009778_050 |
0.1154898488 |
- |
- |
hypothetical protein JCGZ_08917 [Jatropha curcas] |
| 13 |
Hb_001691_060 |
0.1159398948 |
- |
- |
lipoxygenase, putative [Ricinus communis] |
| 14 |
Hb_000029_420 |
0.1298326318 |
- |
- |
PREDICTED: putative lipid-transfer protein DIR1 [Jatropha curcas] |
| 15 |
Hb_006573_260 |
0.130803029 |
- |
- |
- |
| 16 |
Hb_010334_040 |
0.134509083 |
- |
- |
PREDICTED: uncharacterized protein LOC105108579 [Populus euphratica] |
| 17 |
Hb_111985_160 |
0.1351801543 |
- |
- |
hypothetical protein B456_010G008900 [Gossypium raimondii] |
| 18 |
Hb_001377_480 |
0.1385892063 |
transcription factor |
TF Family: MYB |
Myb domain protein 7, putative [Theobroma cacao] |
| 19 |
Hb_001775_050 |
0.1398713463 |
- |
- |
ribosome-inactivating protein cucurmosin-like [Jatropha curcas] |
| 20 |
Hb_001660_040 |
0.1402991796 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Jatropha curcas] |