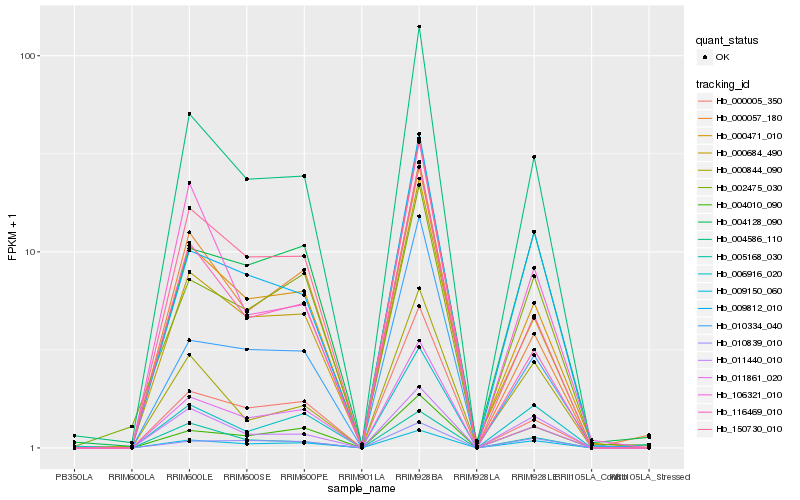
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004586_110 |
0.0 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 2 |
Hb_000471_010 |
0.0540750878 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 3 |
Hb_000684_490 |
0.0570171527 |
transcription factor |
TF Family: WRKY |
PREDICTED: WRKY transcription factor 44 [Jatropha curcas] |
| 4 |
Hb_011861_020 |
0.0892297382 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_06096 [Jatropha curcas] |
| 5 |
Hb_009812_010 |
0.0956838481 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 6 |
Hb_009150_060 |
0.0971230755 |
- |
- |
PREDICTED: cytochrome P450 CYP749A22-like [Prunus mume] |
| 7 |
Hb_005168_030 |
0.1072561702 |
- |
- |
unknown [Zea mays] |
| 8 |
Hb_010334_040 |
0.1102642098 |
- |
- |
PREDICTED: uncharacterized protein LOC105108579 [Populus euphratica] |
| 9 |
Hb_116469_010 |
0.1109363035 |
- |
- |
PREDICTED: uncharacterized protein LOC105647834 [Jatropha curcas] |
| 10 |
Hb_004128_090 |
0.1124249138 |
transcription factor |
TF Family: WRKY |
PREDICTED: WRKY transcription factor 44 [Jatropha curcas] |
| 11 |
Hb_000005_350 |
0.1130305394 |
- |
- |
PREDICTED: uncharacterized protein LOC105634263 [Jatropha curcas] |
| 12 |
Hb_150730_010 |
0.1137742291 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Jatropha curcas] |
| 13 |
Hb_004010_090 |
0.1173177106 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 14 |
Hb_006916_020 |
0.1197135008 |
- |
- |
Flavonol 4'-sulfotransferase, putative [Ricinus communis] |
| 15 |
Hb_000844_090 |
0.1203254232 |
- |
- |
PREDICTED: cytochrome P450 82C4-like [Jatropha curcas] |
| 16 |
Hb_011440_010 |
0.1204902613 |
- |
- |
PREDICTED: metalloendoproteinase 1-like [Pyrus x bretschneideri] |
| 17 |
Hb_000057_180 |
0.1206616905 |
- |
- |
hypothetical protein POPTR_0001s18200g [Populus trichocarpa] |
| 18 |
Hb_010839_010 |
0.1223480224 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 19 |
Hb_002475_030 |
0.1226082738 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_106321_010 |
0.124859678 |
- |
- |
PREDICTED: probable glutathione S-transferase [Jatropha curcas] |