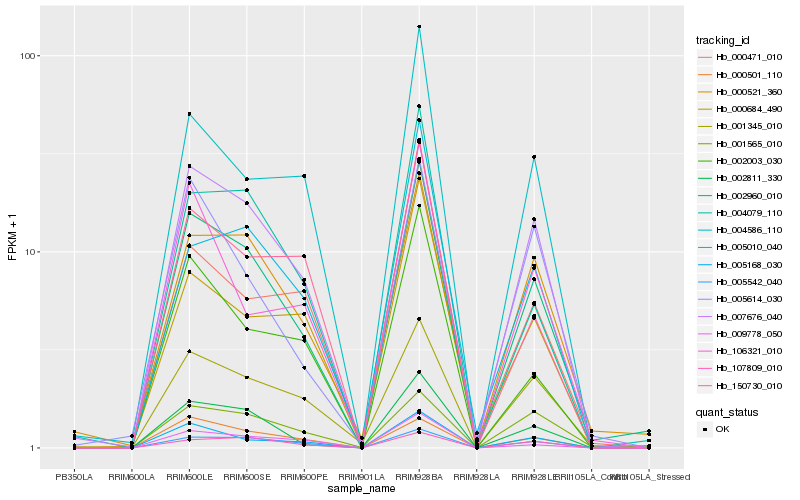
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002960_010 |
0.0 |
- |
- |
Zeamatin precursor, putative [Ricinus communis] |
| 2 |
Hb_002811_330 |
0.1015042854 |
- |
- |
PREDICTED: calmodulin-like protein 11 [Jatropha curcas] |
| 3 |
Hb_000521_360 |
0.1043019851 |
- |
- |
PREDICTED: beta-glucosidase 17-like [Jatropha curcas] |
| 4 |
Hb_007676_040 |
0.107402066 |
- |
- |
cellulose synthase, putative [Ricinus communis] |
| 5 |
Hb_005168_030 |
0.1138908386 |
- |
- |
unknown [Zea mays] |
| 6 |
Hb_005542_040 |
0.1156975328 |
transcription factor |
TF Family: WRKY |
PREDICTED: WRKY transcription factor 55 [Jatropha curcas] |
| 7 |
Hb_001345_010 |
0.1236706507 |
- |
- |
hypothetical protein MIMGU_mgv1a010889mg [Erythranthe guttata] |
| 8 |
Hb_004079_110 |
0.1329584181 |
- |
- |
PREDICTED: uncharacterized protein LOC100257802 [Vitis vinifera] |
| 9 |
Hb_004586_110 |
0.1425997196 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 10 |
Hb_107809_010 |
0.1430056843 |
- |
- |
JHL06P13.14 [Jatropha curcas] |
| 11 |
Hb_002003_030 |
0.144914055 |
- |
- |
PREDICTED: UDP-glycosyltransferase 74F1-like [Jatropha curcas] |
| 12 |
Hb_001565_010 |
0.1451011266 |
- |
- |
hypothetical protein JCGZ_07371 [Jatropha curcas] |
| 13 |
Hb_000501_110 |
0.1468122323 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 [Populus euphratica] |
| 14 |
Hb_000471_010 |
0.1481756023 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 15 |
Hb_000684_490 |
0.1549041466 |
transcription factor |
TF Family: WRKY |
PREDICTED: WRKY transcription factor 44 [Jatropha curcas] |
| 16 |
Hb_150730_010 |
0.1558506494 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Jatropha curcas] |
| 17 |
Hb_005614_030 |
0.1560270252 |
- |
- |
PREDICTED: transcription factor SCREAM2-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_005010_040 |
0.1563899439 |
- |
- |
PREDICTED: uncharacterized protein LOC105630824 [Jatropha curcas] |
| 19 |
Hb_106321_010 |
0.1565164501 |
- |
- |
PREDICTED: probable glutathione S-transferase [Jatropha curcas] |
| 20 |
Hb_009778_050 |
0.1574621473 |
- |
- |
hypothetical protein JCGZ_08917 [Jatropha curcas] |