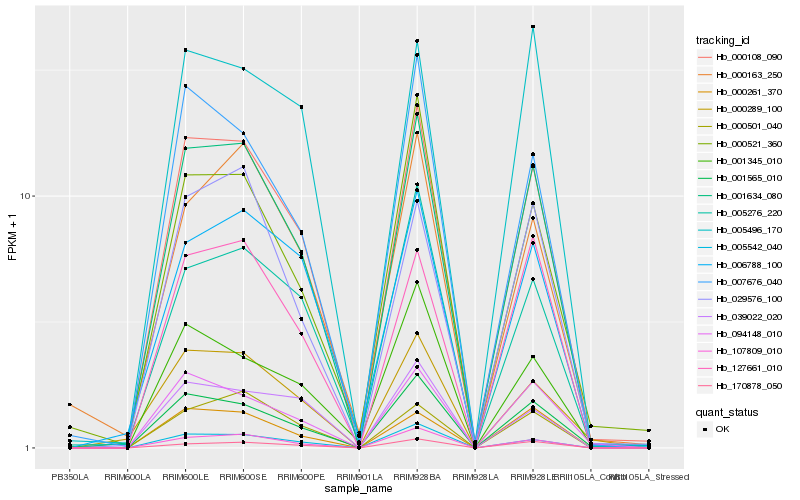
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000108_090 |
0.0 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB5-like [Jatropha curcas] |
| 2 |
Hb_001634_080 |
0.0367750368 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 3 |
Hb_107809_010 |
0.0864466163 |
- |
- |
JHL06P13.14 [Jatropha curcas] |
| 4 |
Hb_001565_010 |
0.0897480405 |
- |
- |
hypothetical protein JCGZ_07371 [Jatropha curcas] |
| 5 |
Hb_005542_040 |
0.0902639222 |
transcription factor |
TF Family: WRKY |
PREDICTED: WRKY transcription factor 55 [Jatropha curcas] |
| 6 |
Hb_007676_040 |
0.0944287989 |
- |
- |
cellulose synthase, putative [Ricinus communis] |
| 7 |
Hb_094148_010 |
0.1042789302 |
- |
- |
PREDICTED: glycerophosphodiester phosphodiesterase protein kinase domain-containing GDPDL2-like isoform X2 [Jatropha curcas] |
| 8 |
Hb_000289_100 |
0.1073413308 |
- |
- |
carbonyl reductase, putative [Ricinus communis] |
| 9 |
Hb_005276_220 |
0.1086297729 |
transcription factor |
TF Family: bHLH |
PREDICTED: basic helix-loop-helix protein A [Jatropha curcas] |
| 10 |
Hb_006788_100 |
0.1094533044 |
- |
- |
PREDICTED: uncharacterized protein LOC105649919 isoform X1 [Jatropha curcas] |
| 11 |
Hb_170878_050 |
0.1125367463 |
- |
- |
retrotransposon protein, putative, unclassified [Oryza sativa Japonica Group] |
| 12 |
Hb_039022_020 |
0.1182800176 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase EFR [Jatropha curcas] |
| 13 |
Hb_127661_010 |
0.1187587085 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 14 |
Hb_000501_040 |
0.1189368985 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 15 |
Hb_000521_360 |
0.1196490127 |
- |
- |
PREDICTED: beta-glucosidase 17-like [Jatropha curcas] |
| 16 |
Hb_000261_370 |
0.1202999903 |
- |
- |
- |
| 17 |
Hb_029576_100 |
0.1212292532 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 31 [Jatropha curcas] |
| 18 |
Hb_001345_010 |
0.123779614 |
- |
- |
hypothetical protein MIMGU_mgv1a010889mg [Erythranthe guttata] |
| 19 |
Hb_000163_250 |
0.1265942784 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 20 |
Hb_005496_170 |
0.1269642424 |
- |
- |
sigma factor sigb regulation protein rsbq, putative [Ricinus communis] |