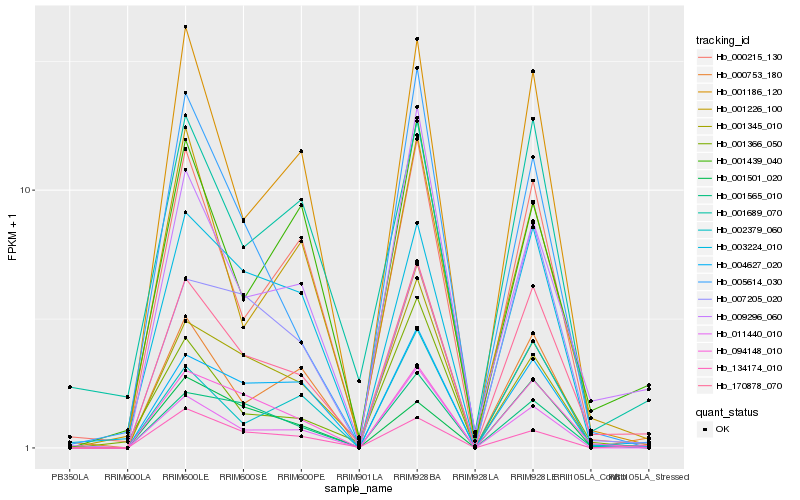
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_170878_070 |
0.0 |
- |
- |
hypothetical protein EUGRSUZ_L008133, partial [Eucalyptus grandis] |
| 2 |
Hb_001565_010 |
0.1100181016 |
- |
- |
hypothetical protein JCGZ_07371 [Jatropha curcas] |
| 3 |
Hb_094148_010 |
0.1156165034 |
- |
- |
PREDICTED: glycerophosphodiester phosphodiesterase protein kinase domain-containing GDPDL2-like isoform X2 [Jatropha curcas] |
| 4 |
Hb_004627_020 |
0.1223959307 |
- |
- |
Protein MLO, putative [Ricinus communis] |
| 5 |
Hb_001186_120 |
0.1375003418 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase 5 [Jatropha curcas] |
| 6 |
Hb_000215_130 |
0.1415150398 |
- |
- |
PREDICTED: probable pectinesterase 53 [Jatropha curcas] |
| 7 |
Hb_001366_050 |
0.1452963399 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription factor NGA1-like isoform X2 [Populus euphratica] |
| 8 |
Hb_011440_010 |
0.1460581103 |
- |
- |
PREDICTED: metalloendoproteinase 1-like [Pyrus x bretschneideri] |
| 9 |
Hb_009296_060 |
0.1464889529 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 10 |
Hb_003224_010 |
0.1471329042 |
- |
- |
- |
| 11 |
Hb_001689_070 |
0.1527199066 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 12 |
Hb_001345_010 |
0.1528929324 |
- |
- |
hypothetical protein MIMGU_mgv1a010889mg [Erythranthe guttata] |
| 13 |
Hb_002379_060 |
0.1538652858 |
- |
- |
PREDICTED: MACPF domain-containing protein At1g14780 [Jatropha curcas] |
| 14 |
Hb_001501_020 |
0.1565259752 |
- |
- |
PREDICTED: receptor-like protein kinase [Jatropha curcas] |
| 15 |
Hb_134174_010 |
0.1578592794 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 16 |
Hb_001439_040 |
0.1615455787 |
- |
- |
PREDICTED: chorismate mutase 1, chloroplastic [Jatropha curcas] |
| 17 |
Hb_001226_100 |
0.1617682611 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 18 |
Hb_007205_020 |
0.1626443218 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Jatropha curcas] |
| 19 |
Hb_000753_180 |
0.1627892554 |
- |
- |
PREDICTED: dirigent protein 22-like [Jatropha curcas] |
| 20 |
Hb_005614_030 |
0.1655173826 |
- |
- |
PREDICTED: transcription factor SCREAM2-like isoform X1 [Jatropha curcas] |