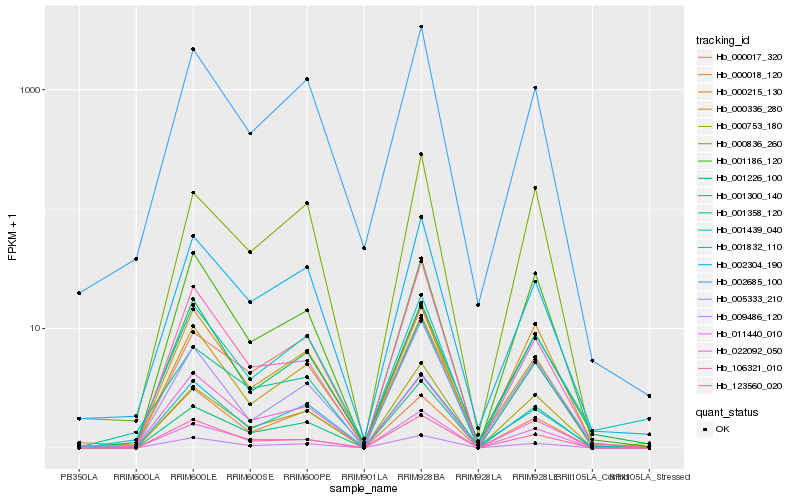
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000336_280 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632845 [Jatropha curcas] |
| 2 |
Hb_001832_110 |
0.0576279191 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase ALE2 isoform X1 [Jatropha curcas] |
| 3 |
Hb_002685_100 |
0.0711543958 |
- |
- |
low temperature inducible SLTI66 [Glycine max] |
| 4 |
Hb_001226_100 |
0.0852653033 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 5 |
Hb_005333_210 |
0.0880859714 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH30-like [Jatropha curcas] |
| 6 |
Hb_000215_130 |
0.0907941709 |
- |
- |
PREDICTED: probable pectinesterase 53 [Jatropha curcas] |
| 7 |
Hb_002304_190 |
0.0932256886 |
- |
- |
PREDICTED: 3'-N-debenzoyl-2'-deoxytaxol N-benzoyltransferase [Jatropha curcas] |
| 8 |
Hb_123560_020 |
0.1048080807 |
- |
- |
hypothetical protein JCGZ_12464 [Jatropha curcas] |
| 9 |
Hb_009486_120 |
0.1069574018 |
- |
- |
hypothetical protein PRUPE_ppa004149mg [Prunus persica] |
| 10 |
Hb_001439_040 |
0.1140471027 |
- |
- |
PREDICTED: chorismate mutase 1, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000018_120 |
0.1171006138 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 12 |
Hb_001186_120 |
0.1182168944 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase 5 [Jatropha curcas] |
| 13 |
Hb_000753_180 |
0.1251933012 |
- |
- |
PREDICTED: dirigent protein 22-like [Jatropha curcas] |
| 14 |
Hb_000836_260 |
0.1269775292 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 15 |
Hb_001300_140 |
0.1284889763 |
- |
- |
- |
| 16 |
Hb_001358_120 |
0.1296731819 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 308-like [Populus euphratica] |
| 17 |
Hb_011440_010 |
0.1310726768 |
- |
- |
PREDICTED: metalloendoproteinase 1-like [Pyrus x bretschneideri] |
| 18 |
Hb_022092_050 |
0.1401295093 |
- |
- |
PREDICTED: uncharacterized protein LOC105136214 [Populus euphratica] |
| 19 |
Hb_000017_320 |
0.1440896676 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor GLABRA 3-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_106321_010 |
0.1481890734 |
- |
- |
PREDICTED: probable glutathione S-transferase [Jatropha curcas] |