| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
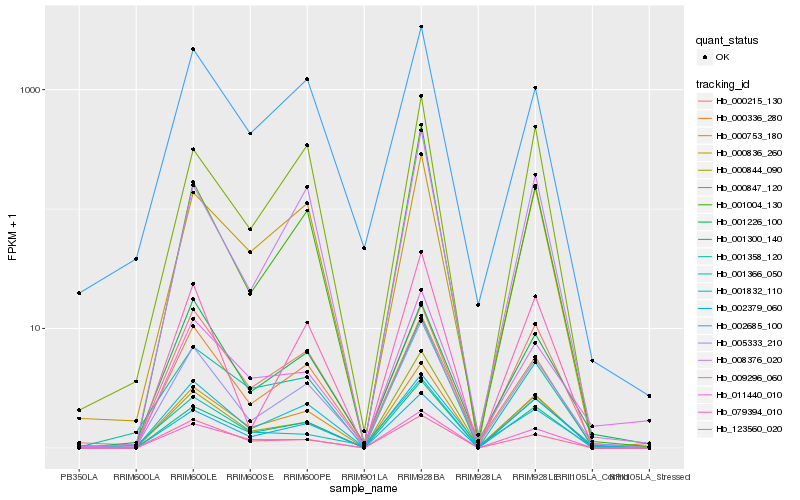
Hb_005333_210 |
0.0 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH30-like [Jatropha curcas] |
| 2 |
Hb_000336_280 |
0.0880859714 |
- |
- |
PREDICTED: uncharacterized protein LOC105632845 [Jatropha curcas] |
| 3 |
Hb_001300_140 |
0.1011318069 |
- |
- |
- |
| 4 |
Hb_079394_010 |
0.1093177962 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 5 |
Hb_000215_130 |
0.1130067219 |
- |
- |
PREDICTED: probable pectinesterase 53 [Jatropha curcas] |
| 6 |
Hb_000753_180 |
0.114460943 |
- |
- |
PREDICTED: dirigent protein 22-like [Jatropha curcas] |
| 7 |
Hb_001004_130 |
0.114481346 |
- |
- |
PREDICTED: chalcone synthase 3 [Jatropha curcas] |
| 8 |
Hb_002685_100 |
0.1167482854 |
- |
- |
low temperature inducible SLTI66 [Glycine max] |
| 9 |
Hb_011440_010 |
0.1169076476 |
- |
- |
PREDICTED: metalloendoproteinase 1-like [Pyrus x bretschneideri] |
| 10 |
Hb_000844_090 |
0.11718201 |
- |
- |
PREDICTED: cytochrome P450 82C4-like [Jatropha curcas] |
| 11 |
Hb_000836_260 |
0.1216428931 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 12 |
Hb_001226_100 |
0.1221672841 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 13 |
Hb_008376_020 |
0.1243928393 |
- |
- |
hypothetical protein JCGZ_03321 [Jatropha curcas] |
| 14 |
Hb_001832_110 |
0.1244086942 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase ALE2 isoform X1 [Jatropha curcas] |
| 15 |
Hb_009296_060 |
0.1333325092 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 16 |
Hb_001358_120 |
0.1336923289 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 308-like [Populus euphratica] |
| 17 |
Hb_123560_020 |
0.1360942105 |
- |
- |
hypothetical protein JCGZ_12464 [Jatropha curcas] |
| 18 |
Hb_002379_060 |
0.1385690006 |
- |
- |
PREDICTED: MACPF domain-containing protein At1g14780 [Jatropha curcas] |
| 19 |
Hb_001366_050 |
0.1394825345 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription factor NGA1-like isoform X2 [Populus euphratica] |
| 20 |
Hb_000847_120 |
0.1411869778 |
- |
- |
hypothetical protein JCGZ_15115 [Jatropha curcas] |