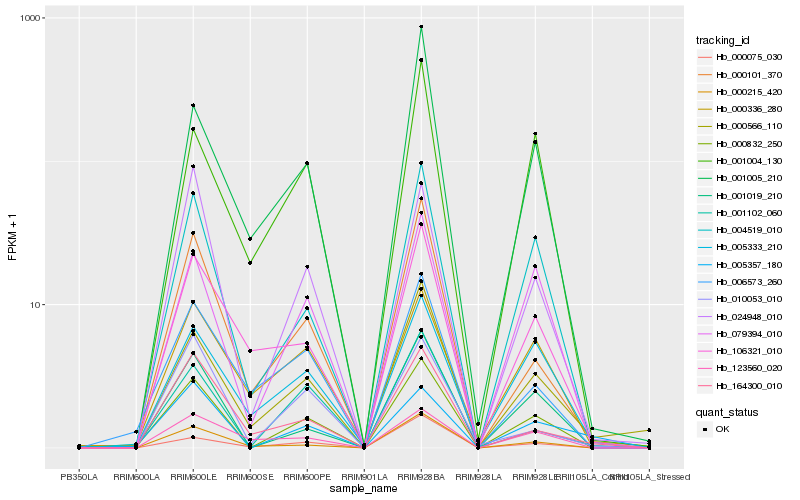
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000832_250 |
0.0 |
- |
- |
PREDICTED: protein UPSTREAM OF FLC-like [Jatropha curcas] |
| 2 |
Hb_004519_010 |
0.1178921864 |
- |
- |
PREDICTED: basic form of pathogenesis-related protein 1-like [Jatropha curcas] |
| 3 |
Hb_001019_210 |
0.1248093659 |
- |
- |
PREDICTED: protein kinase PINOID 2 [Gossypium raimondii] |
| 4 |
Hb_079394_010 |
0.1297937294 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 5 |
Hb_024948_010 |
0.1537152112 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase, basic isoform-like [Populus euphratica] |
| 6 |
Hb_000215_420 |
0.1610312975 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor RAX2-like [Populus euphratica] |
| 7 |
Hb_000101_370 |
0.1615191043 |
- |
- |
PREDICTED: dihydrofolate reductase-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_000566_110 |
0.1647290855 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 75 [Prunus mume] |
| 9 |
Hb_001102_060 |
0.1658568384 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase At1g78530 isoform X1 [Jatropha curcas] |
| 10 |
Hb_006573_260 |
0.1743899048 |
- |
- |
- |
| 11 |
Hb_005333_210 |
0.176364381 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH30-like [Jatropha curcas] |
| 12 |
Hb_106321_010 |
0.176436906 |
- |
- |
PREDICTED: probable glutathione S-transferase [Jatropha curcas] |
| 13 |
Hb_001004_130 |
0.1782054328 |
- |
- |
PREDICTED: chalcone synthase 3 [Jatropha curcas] |
| 14 |
Hb_005357_180 |
0.182376778 |
- |
- |
hypothetical protein POPTR_0011s00490g [Populus trichocarpa] |
| 15 |
Hb_001005_210 |
0.1852757535 |
- |
- |
PREDICTED: chalcone synthase 3 [Jatropha curcas] |
| 16 |
Hb_000336_280 |
0.1869307104 |
- |
- |
PREDICTED: uncharacterized protein LOC105632845 [Jatropha curcas] |
| 17 |
Hb_010053_010 |
0.1880765037 |
- |
- |
hypothetical protein POPTR_0515s00220g [Populus trichocarpa] |
| 18 |
Hb_000075_030 |
0.190383891 |
- |
- |
PREDICTED: aldo-keto reductase family 4 member C10-like [Fragaria vesca subsp. vesca] |
| 19 |
Hb_164300_010 |
0.1923715385 |
- |
- |
PREDICTED: thaumatin-like protein 1b [Jatropha curcas] |
| 20 |
Hb_123560_020 |
0.1970914458 |
- |
- |
hypothetical protein JCGZ_12464 [Jatropha curcas] |