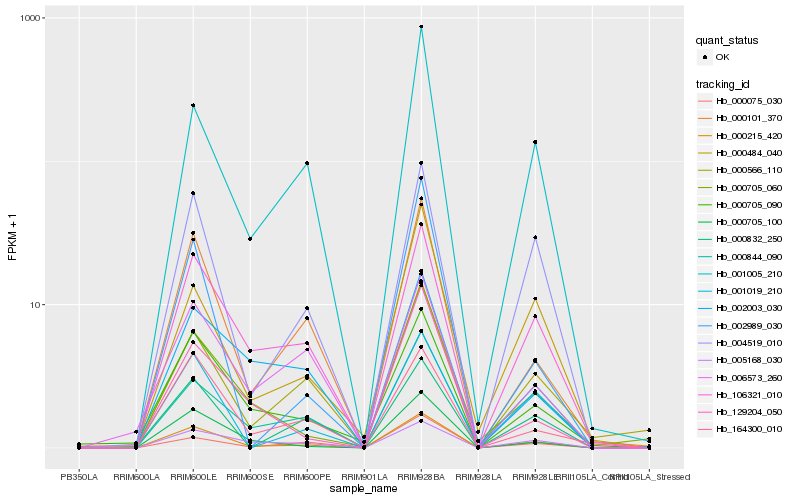
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000215_420 |
0.0 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor RAX2-like [Populus euphratica] |
| 2 |
Hb_106321_010 |
0.1070459436 |
- |
- |
PREDICTED: probable glutathione S-transferase [Jatropha curcas] |
| 3 |
Hb_000101_370 |
0.1145596765 |
- |
- |
PREDICTED: dihydrofolate reductase-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_004519_010 |
0.1145622108 |
- |
- |
PREDICTED: basic form of pathogenesis-related protein 1-like [Jatropha curcas] |
| 5 |
Hb_000705_100 |
0.1236539138 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 6 |
Hb_000705_090 |
0.1264844499 |
- |
- |
PREDICTED: uncharacterized protein LOC100246830 [Vitis vinifera] |
| 7 |
Hb_001019_210 |
0.1373170499 |
- |
- |
PREDICTED: protein kinase PINOID 2 [Gossypium raimondii] |
| 8 |
Hb_001005_210 |
0.1393385589 |
- |
- |
PREDICTED: chalcone synthase 3 [Jatropha curcas] |
| 9 |
Hb_000705_060 |
0.1412484551 |
- |
- |
hypothetical protein TRIUR3_06087 [Triticum urartu] |
| 10 |
Hb_164300_010 |
0.1481106794 |
- |
- |
PREDICTED: thaumatin-like protein 1b [Jatropha curcas] |
| 11 |
Hb_000566_110 |
0.1528563994 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 75 [Prunus mume] |
| 12 |
Hb_005168_030 |
0.1542609769 |
- |
- |
unknown [Zea mays] |
| 13 |
Hb_000075_030 |
0.1604765232 |
- |
- |
PREDICTED: aldo-keto reductase family 4 member C10-like [Fragaria vesca subsp. vesca] |
| 14 |
Hb_129204_050 |
0.1606252653 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-related protein 330-like [Prunus mume] |
| 15 |
Hb_000832_250 |
0.1610312975 |
- |
- |
PREDICTED: protein UPSTREAM OF FLC-like [Jatropha curcas] |
| 16 |
Hb_000484_040 |
0.1654469013 |
- |
- |
PREDICTED: zeaxanthin epoxidase, chloroplastic-like [Jatropha curcas] |
| 17 |
Hb_000844_090 |
0.1677867078 |
- |
- |
PREDICTED: cytochrome P450 82C4-like [Jatropha curcas] |
| 18 |
Hb_006573_260 |
0.1715795757 |
- |
- |
- |
| 19 |
Hb_002989_030 |
0.1727986469 |
- |
- |
PREDICTED: protein GAST1-like [Populus euphratica] |
| 20 |
Hb_002003_030 |
0.173759685 |
- |
- |
PREDICTED: UDP-glycosyltransferase 74F1-like [Jatropha curcas] |