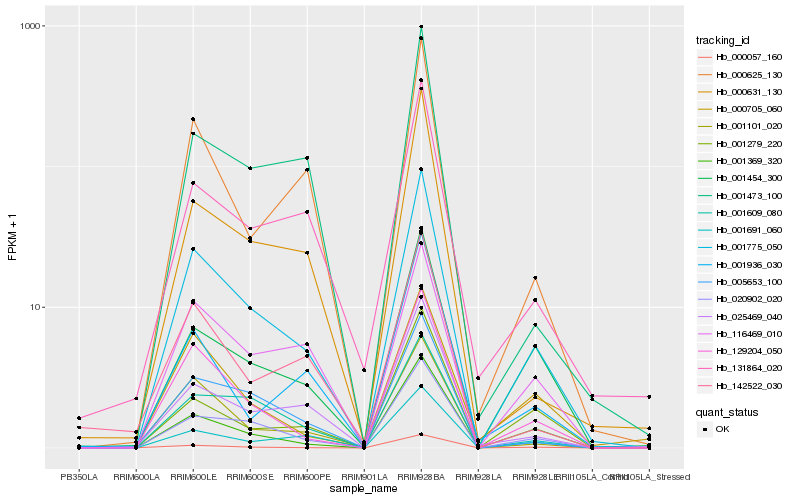
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001775_050 |
0.0 |
- |
- |
ribosome-inactivating protein cucurmosin-like [Jatropha curcas] |
| 2 |
Hb_020902_020 |
0.0849139082 |
- |
- |
hypothetical protein JCGZ_07820 [Jatropha curcas] |
| 3 |
Hb_129204_050 |
0.0868891794 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-related protein 330-like [Prunus mume] |
| 4 |
Hb_001369_320 |
0.1045146734 |
- |
- |
PREDICTED: dynein light chain, cytoplasmic-like [Jatropha curcas] |
| 5 |
Hb_000057_160 |
0.112244667 |
- |
- |
hypothetical protein JCGZ_06279 [Jatropha curcas] |
| 6 |
Hb_005653_100 |
0.1203279295 |
- |
- |
hypothetical protein POPTR_0004s19580g [Populus trichocarpa] |
| 7 |
Hb_001454_300 |
0.1215931371 |
- |
- |
nucleotide binding protein, putative [Ricinus communis] |
| 8 |
Hb_001101_020 |
0.1257085471 |
- |
- |
hypothetical protein RCOM_0615680 [Ricinus communis] |
| 9 |
Hb_025469_040 |
0.1306273238 |
- |
- |
PREDICTED: acidic mammalian chitinase-like [Jatropha curcas] |
| 10 |
Hb_001691_060 |
0.1359762119 |
- |
- |
lipoxygenase, putative [Ricinus communis] |
| 11 |
Hb_001609_080 |
0.1371470573 |
- |
- |
hypothetical protein POPTR_0017s05660g [Populus trichocarpa] |
| 12 |
Hb_000625_130 |
0.1383320573 |
- |
- |
PREDICTED: naringenin,2-oxoglutarate 3-dioxygenase [Jatropha curcas] |
| 13 |
Hb_116469_010 |
0.1398713463 |
- |
- |
PREDICTED: uncharacterized protein LOC105647834 [Jatropha curcas] |
| 14 |
Hb_000631_130 |
0.140128565 |
- |
- |
PREDICTED: flavonoid 3'-monooxygenase [Jatropha curcas] |
| 15 |
Hb_001279_220 |
0.1423826084 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 16 |
Hb_131864_020 |
0.1434517731 |
- |
- |
PREDICTED: anthocyanidin reductase [Jatropha curcas] |
| 17 |
Hb_000705_060 |
0.146342528 |
- |
- |
hypothetical protein TRIUR3_06087 [Triticum urartu] |
| 18 |
Hb_001473_100 |
0.1513060291 |
- |
- |
chalcone synthase 1 [Jatropha curcas] |
| 19 |
Hb_142522_030 |
0.1524390001 |
- |
- |
hypothetical protein JCGZ_21596 [Jatropha curcas] |
| 20 |
Hb_001936_030 |
0.1524647651 |
- |
- |
PREDICTED: polygalacturonase At1g48100 isoform X1 [Jatropha curcas] |