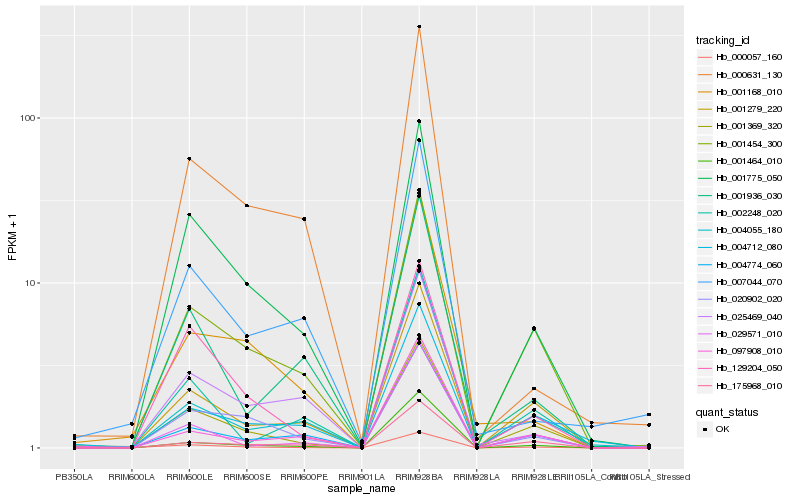
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000057_160 |
0.0 |
- |
- |
hypothetical protein JCGZ_06279 [Jatropha curcas] |
| 2 |
Hb_001369_320 |
0.1012592542 |
- |
- |
PREDICTED: dynein light chain, cytoplasmic-like [Jatropha curcas] |
| 3 |
Hb_001775_050 |
0.112244667 |
- |
- |
ribosome-inactivating protein cucurmosin-like [Jatropha curcas] |
| 4 |
Hb_001279_220 |
0.1232381793 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 5 |
Hb_001454_300 |
0.1326392691 |
- |
- |
nucleotide binding protein, putative [Ricinus communis] |
| 6 |
Hb_129204_050 |
0.1349139262 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-related protein 330-like [Prunus mume] |
| 7 |
Hb_029571_010 |
0.1383638119 |
- |
- |
PREDICTED: polygalacturonase QRT3 [Jatropha curcas] |
| 8 |
Hb_004774_060 |
0.1389674522 |
- |
- |
- |
| 9 |
Hb_020902_020 |
0.1427078281 |
- |
- |
hypothetical protein JCGZ_07820 [Jatropha curcas] |
| 10 |
Hb_001464_010 |
0.1460230706 |
- |
- |
PREDICTED: putative receptor-like protein kinase At4g00960 isoform X1 [Populus euphratica] |
| 11 |
Hb_001168_010 |
0.1472502365 |
- |
- |
PREDICTED: feruloyl CoA ortho-hydroxylase 1 [Jatropha curcas] |
| 12 |
Hb_175968_010 |
0.1509686458 |
- |
- |
hypothetical protein JCGZ_21281 [Jatropha curcas] |
| 13 |
Hb_002248_020 |
0.1514961972 |
transcription factor |
TF Family: NAC |
PREDICTED: transcription factor JUNGBRUNNEN 1-like isoform X1 [Populus euphratica] |
| 14 |
Hb_004055_180 |
0.1526487061 |
- |
- |
cell wall invertase [Manihot esculenta] |
| 15 |
Hb_001936_030 |
0.1529152776 |
- |
- |
PREDICTED: polygalacturonase At1g48100 isoform X1 [Jatropha curcas] |
| 16 |
Hb_025469_040 |
0.1565850796 |
- |
- |
PREDICTED: acidic mammalian chitinase-like [Jatropha curcas] |
| 17 |
Hb_004712_080 |
0.1631032748 |
- |
- |
PREDICTED: premnaspirodiene oxygenase-like [Jatropha curcas] |
| 18 |
Hb_000631_130 |
0.1656391798 |
- |
- |
PREDICTED: flavonoid 3'-monooxygenase [Jatropha curcas] |
| 19 |
Hb_007044_070 |
0.1657659494 |
- |
- |
PREDICTED: tetraketide alpha-pyrone reductase 1-like [Jatropha curcas] |
| 20 |
Hb_097908_010 |
0.1658733135 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g03230 isoform X2 [Jatropha curcas] |