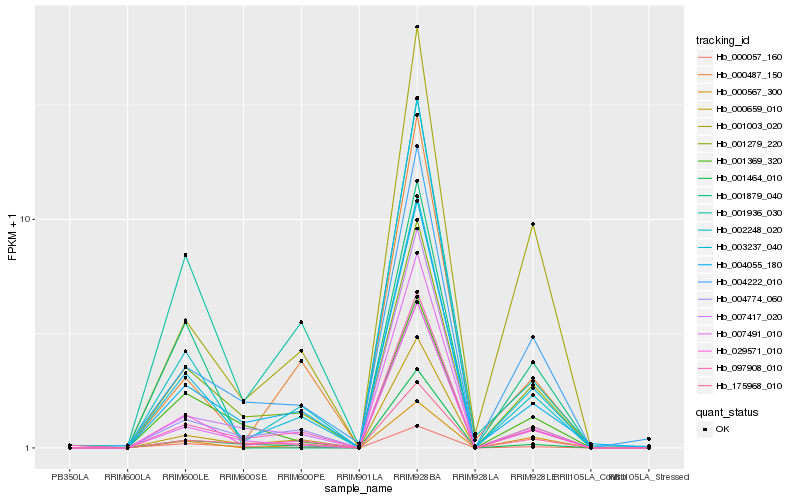
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_029571_010 |
0.0 |
- |
- |
PREDICTED: polygalacturonase QRT3 [Jatropha curcas] |
| 2 |
Hb_002248_020 |
0.0537124962 |
transcription factor |
TF Family: NAC |
PREDICTED: transcription factor JUNGBRUNNEN 1-like isoform X1 [Populus euphratica] |
| 3 |
Hb_004055_180 |
0.0997814807 |
- |
- |
cell wall invertase [Manihot esculenta] |
| 4 |
Hb_004774_060 |
0.1064544231 |
- |
- |
- |
| 5 |
Hb_001279_220 |
0.1067932848 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 6 |
Hb_001464_010 |
0.1097121361 |
- |
- |
PREDICTED: putative receptor-like protein kinase At4g00960 isoform X1 [Populus euphratica] |
| 7 |
Hb_097908_010 |
0.1114717659 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g03230 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000659_010 |
0.1141630728 |
- |
- |
PREDICTED: plasma membrane ATPase 2-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_004222_010 |
0.1267937019 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal-like [Jatropha curcas] |
| 10 |
Hb_007417_020 |
0.1269957453 |
- |
- |
Reticuline oxidase precursor, putative [Ricinus communis] |
| 11 |
Hb_175968_010 |
0.1277512323 |
- |
- |
hypothetical protein JCGZ_21281 [Jatropha curcas] |
| 12 |
Hb_001879_040 |
0.1351445766 |
- |
- |
hypothetical protein CISIN_1g0033281mg, partial [Citrus sinensis] |
| 13 |
Hb_007491_010 |
0.1372410459 |
- |
- |
Spotted leaf protein, putative [Ricinus communis] |
| 14 |
Hb_001936_030 |
0.1375768719 |
- |
- |
PREDICTED: polygalacturonase At1g48100 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000057_160 |
0.1383638119 |
- |
- |
hypothetical protein JCGZ_06279 [Jatropha curcas] |
| 16 |
Hb_003237_040 |
0.1429868486 |
- |
- |
Ent-kaurene synthase B, chloroplast precursor, putative [Ricinus communis] |
| 17 |
Hb_000487_150 |
0.1443286036 |
- |
- |
PREDICTED: feruloyl CoA ortho-hydroxylase 2-like [Jatropha curcas] |
| 18 |
Hb_001003_020 |
0.1449087627 |
- |
- |
PREDICTED: metalloendoproteinase 1-like [Populus euphratica] |
| 19 |
Hb_001369_320 |
0.150594001 |
- |
- |
PREDICTED: dynein light chain, cytoplasmic-like [Jatropha curcas] |
| 20 |
Hb_000567_300 |
0.1536644987 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |