| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
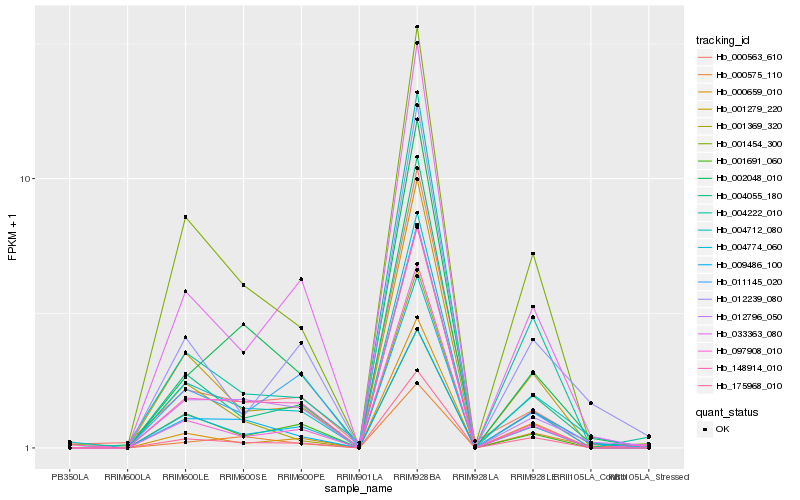
Hb_004712_080 |
0.0 |
- |
- |
PREDICTED: premnaspirodiene oxygenase-like [Jatropha curcas] |
| 2 |
Hb_012796_050 |
0.1023673929 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
1-deoxyxylulose-5-phosphate synthase, putative [Ricinus communis] |
| 3 |
Hb_175968_010 |
0.1032432596 |
- |
- |
hypothetical protein JCGZ_21281 [Jatropha curcas] |
| 4 |
Hb_001279_220 |
0.1047575817 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 5 |
Hb_033363_080 |
0.1067551817 |
- |
- |
Allene oxide cyclase 3, chloroplast precursor, putative [Ricinus communis] |
| 6 |
Hb_001454_300 |
0.1091076469 |
- |
- |
nucleotide binding protein, putative [Ricinus communis] |
| 7 |
Hb_004774_060 |
0.1105150958 |
- |
- |
- |
| 8 |
Hb_004055_180 |
0.1185777226 |
- |
- |
cell wall invertase [Manihot esculenta] |
| 9 |
Hb_097908_010 |
0.1191647916 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g03230 isoform X2 [Jatropha curcas] |
| 10 |
Hb_012239_080 |
0.1238606956 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase homolog 4-like [Jatropha curcas] |
| 11 |
Hb_002048_010 |
0.1334511293 |
- |
- |
unknown [Lotus japonicus] |
| 12 |
Hb_000563_610 |
0.1334955998 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase LIN-1 [Jatropha curcas] |
| 13 |
Hb_148914_010 |
0.1366712596 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 14 |
Hb_000659_010 |
0.1422686769 |
- |
- |
PREDICTED: plasma membrane ATPase 2-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_001691_060 |
0.1426659869 |
- |
- |
lipoxygenase, putative [Ricinus communis] |
| 16 |
Hb_001369_320 |
0.1453877215 |
- |
- |
PREDICTED: dynein light chain, cytoplasmic-like [Jatropha curcas] |
| 17 |
Hb_009486_100 |
0.1462078382 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 18 |
Hb_000575_110 |
0.1470116534 |
- |
- |
S-locus-specific glycoprotein S13 precursor, putative [Ricinus communis] |
| 19 |
Hb_011145_020 |
0.148607588 |
- |
- |
PREDICTED: metalloendoproteinase 1-like [Pyrus x bretschneideri] |
| 20 |
Hb_004222_010 |
0.1487647014 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal-like [Jatropha curcas] |