| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
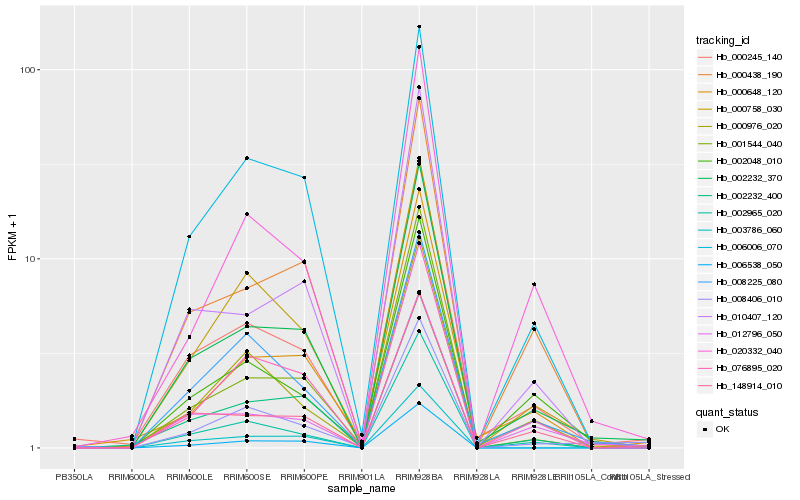
Hb_002232_370 |
0.0 |
- |
- |
PREDICTED: receptor-like protein kinase HAIKU2 [Jatropha curcas] |
| 2 |
Hb_000245_140 |
0.0667845991 |
- |
- |
PREDICTED: CASP-like protein 3A1 [Jatropha curcas] |
| 3 |
Hb_000438_190 |
0.085133123 |
- |
- |
PREDICTED: peroxidase 4-like [Jatropha curcas] |
| 4 |
Hb_148914_010 |
0.0881725665 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 5 |
Hb_002965_020 |
0.0904431902 |
- |
- |
hypothetical protein JCGZ_18947 [Jatropha curcas] |
| 6 |
Hb_002232_400 |
0.0946936885 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH25-like [Jatropha curcas] |
| 7 |
Hb_012796_050 |
0.0958365071 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
1-deoxyxylulose-5-phosphate synthase, putative [Ricinus communis] |
| 8 |
Hb_006006_070 |
0.1005024023 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 9 |
Hb_010407_120 |
0.100596648 |
- |
- |
PREDICTED: glutathione transferase GST 23-like [Jatropha curcas] |
| 10 |
Hb_001544_040 |
0.103716532 |
- |
- |
PREDICTED: uncharacterized protein At1g66480-like [Populus euphratica] |
| 11 |
Hb_076895_020 |
0.1088423353 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000648_120 |
0.1107522032 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase IV.1 [Vitis vinifera] |
| 13 |
Hb_006538_050 |
0.1111688641 |
- |
- |
hypothetical protein RCOM_1308310 [Ricinus communis] |
| 14 |
Hb_000758_030 |
0.1119594204 |
transcription factor |
TF Family: ERF |
Ethylene-responsive transcription factor, putative [Ricinus communis] |
| 15 |
Hb_003786_060 |
0.1125445405 |
- |
- |
PREDICTED: transmembrane protein 220 [Jatropha curcas] |
| 16 |
Hb_008406_010 |
0.1126094174 |
- |
- |
PREDICTED: tyrosine aminotransferase-like [Populus euphratica] |
| 17 |
Hb_002048_010 |
0.1143489288 |
- |
- |
unknown [Lotus japonicus] |
| 18 |
Hb_008225_080 |
0.1167975592 |
- |
- |
PREDICTED: cytochrome P450 71A26-like [Jatropha curcas] |
| 19 |
Hb_020332_040 |
0.118853712 |
- |
- |
PREDICTED: alcohol dehydrogenase 1-like [Jatropha curcas] |
| 20 |
Hb_000976_020 |
0.1197219267 |
- |
- |
leucoanthocyanidin dioxygenase, putative [Ricinus communis] |