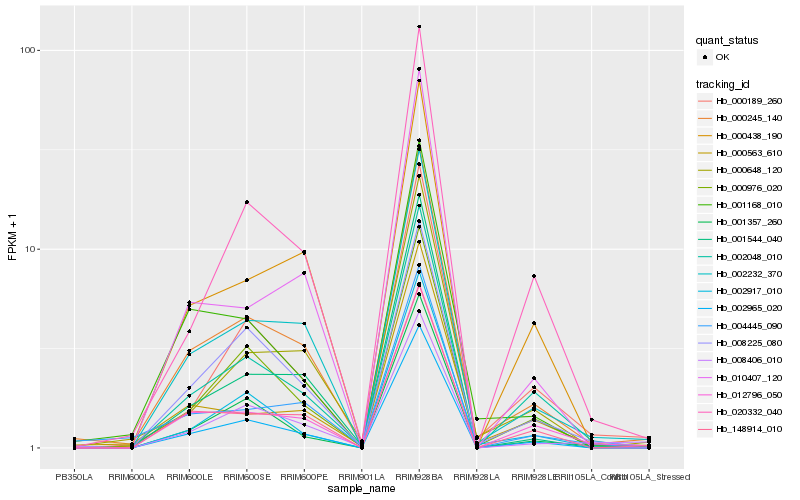
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000245_140 |
0.0 |
- |
- |
PREDICTED: CASP-like protein 3A1 [Jatropha curcas] |
| 2 |
Hb_002232_370 |
0.0667845991 |
- |
- |
PREDICTED: receptor-like protein kinase HAIKU2 [Jatropha curcas] |
| 3 |
Hb_002965_020 |
0.0821565628 |
- |
- |
hypothetical protein JCGZ_18947 [Jatropha curcas] |
| 4 |
Hb_148914_010 |
0.0951501481 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 5 |
Hb_000976_020 |
0.0982380385 |
- |
- |
leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 6 |
Hb_000648_120 |
0.1001274417 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase IV.1 [Vitis vinifera] |
| 7 |
Hb_000563_610 |
0.1009922203 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase LIN-1 [Jatropha curcas] |
| 8 |
Hb_012796_050 |
0.105530015 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
1-deoxyxylulose-5-phosphate synthase, putative [Ricinus communis] |
| 9 |
Hb_002048_010 |
0.1087164002 |
- |
- |
unknown [Lotus japonicus] |
| 10 |
Hb_000438_190 |
0.1088611468 |
- |
- |
PREDICTED: peroxidase 4-like [Jatropha curcas] |
| 11 |
Hb_001544_040 |
0.1095965873 |
- |
- |
PREDICTED: uncharacterized protein At1g66480-like [Populus euphratica] |
| 12 |
Hb_001168_010 |
0.1116717986 |
- |
- |
PREDICTED: feruloyl CoA ortho-hydroxylase 1 [Jatropha curcas] |
| 13 |
Hb_008225_080 |
0.1117375531 |
- |
- |
PREDICTED: cytochrome P450 71A26-like [Jatropha curcas] |
| 14 |
Hb_004445_090 |
0.1117618445 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_008406_010 |
0.1139322259 |
- |
- |
PREDICTED: tyrosine aminotransferase-like [Populus euphratica] |
| 16 |
Hb_010407_120 |
0.1139745234 |
- |
- |
PREDICTED: glutathione transferase GST 23-like [Jatropha curcas] |
| 17 |
Hb_001357_260 |
0.1153779434 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 18 |
Hb_020332_040 |
0.1155740559 |
- |
- |
PREDICTED: alcohol dehydrogenase 1-like [Jatropha curcas] |
| 19 |
Hb_002917_010 |
0.1158985999 |
- |
- |
PREDICTED: receptor-like protein kinase HSL1 [Jatropha curcas] |
| 20 |
Hb_000189_260 |
0.1173357108 |
- |
- |
conserved hypothetical protein [Ricinus communis] |