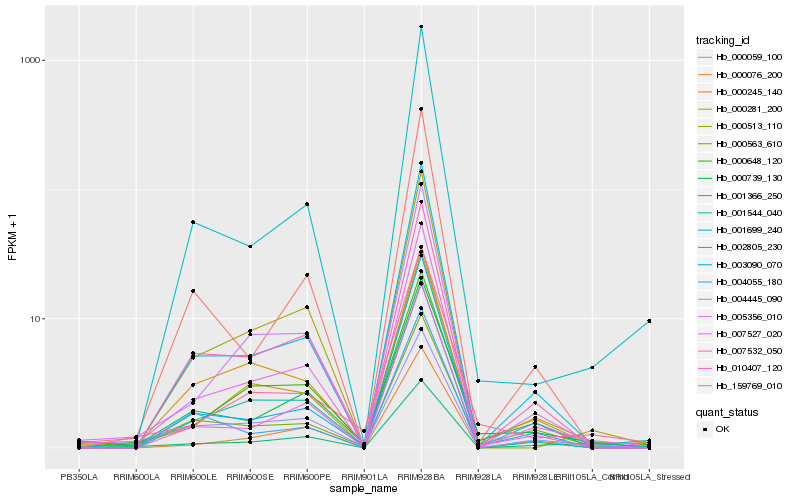
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000739_130 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000563_610 |
0.0983637337 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase LIN-1 [Jatropha curcas] |
| 3 |
Hb_007532_050 |
0.1134131361 |
- |
- |
9-cis-epoxycarotenoid dioxygenase, putative [Ricinus communis] |
| 4 |
Hb_007527_020 |
0.1144645242 |
- |
- |
PREDICTED: feruloyl CoA ortho-hydroxylase 1 [Jatropha curcas] |
| 5 |
Hb_010407_120 |
0.1219289462 |
- |
- |
PREDICTED: glutathione transferase GST 23-like [Jatropha curcas] |
| 6 |
Hb_004445_090 |
0.1262768775 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_159769_010 |
0.1334672624 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 8 |
Hb_000648_120 |
0.1352387995 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase IV.1 [Vitis vinifera] |
| 9 |
Hb_003090_070 |
0.1378582811 |
- |
- |
PREDICTED: laccase-7 [Jatropha curcas] |
| 10 |
Hb_000281_200 |
0.138268125 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 [Jatropha curcas] |
| 11 |
Hb_000059_100 |
0.1393156796 |
- |
- |
Basic blue protein, putative [Ricinus communis] |
| 12 |
Hb_000245_140 |
0.1393297419 |
- |
- |
PREDICTED: CASP-like protein 3A1 [Jatropha curcas] |
| 13 |
Hb_002805_230 |
0.1409305444 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001544_040 |
0.1416262004 |
- |
- |
PREDICTED: uncharacterized protein At1g66480-like [Populus euphratica] |
| 15 |
Hb_000513_110 |
0.1456207164 |
- |
- |
PREDICTED: germin-like protein subfamily 1 member 13 [Populus euphratica] |
| 16 |
Hb_005356_010 |
0.1468559757 |
- |
- |
hypothetical protein POPTR_0003s16550g [Populus trichocarpa] |
| 17 |
Hb_000076_200 |
0.1470109783 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_004055_180 |
0.1485820952 |
- |
- |
cell wall invertase [Manihot esculenta] |
| 19 |
Hb_001366_250 |
0.1491393951 |
- |
- |
PREDICTED: mannan endo-1,4-beta-mannosidase 4-like [Jatropha curcas] |
| 20 |
Hb_001699_240 |
0.149651266 |
- |
- |
PREDICTED: naringenin,2-oxoglutarate 3-dioxygenase [Jatropha curcas] |