| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
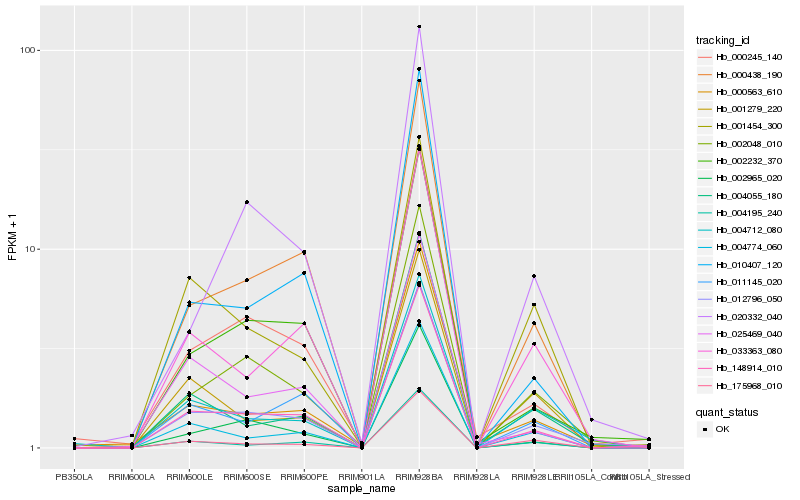
Hb_012796_050 |
0.0 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
1-deoxyxylulose-5-phosphate synthase, putative [Ricinus communis] |
| 2 |
Hb_148914_010 |
0.0636740525 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 3 |
Hb_002048_010 |
0.0818716713 |
- |
- |
unknown [Lotus japonicus] |
| 4 |
Hb_002232_370 |
0.0958365071 |
- |
- |
PREDICTED: receptor-like protein kinase HAIKU2 [Jatropha curcas] |
| 5 |
Hb_000438_190 |
0.0972835764 |
- |
- |
PREDICTED: peroxidase 4-like [Jatropha curcas] |
| 6 |
Hb_004712_080 |
0.1023673929 |
- |
- |
PREDICTED: premnaspirodiene oxygenase-like [Jatropha curcas] |
| 7 |
Hb_033363_080 |
0.1026080749 |
- |
- |
Allene oxide cyclase 3, chloroplast precursor, putative [Ricinus communis] |
| 8 |
Hb_000245_140 |
0.105530015 |
- |
- |
PREDICTED: CASP-like protein 3A1 [Jatropha curcas] |
| 9 |
Hb_004774_060 |
0.1095955312 |
- |
- |
- |
| 10 |
Hb_002965_020 |
0.1103140206 |
- |
- |
hypothetical protein JCGZ_18947 [Jatropha curcas] |
| 11 |
Hb_000563_610 |
0.1116004794 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase LIN-1 [Jatropha curcas] |
| 12 |
Hb_175968_010 |
0.1183234936 |
- |
- |
hypothetical protein JCGZ_21281 [Jatropha curcas] |
| 13 |
Hb_004055_180 |
0.1185446787 |
- |
- |
cell wall invertase [Manihot esculenta] |
| 14 |
Hb_011145_020 |
0.1243432407 |
- |
- |
PREDICTED: metalloendoproteinase 1-like [Pyrus x bretschneideri] |
| 15 |
Hb_010407_120 |
0.1256301171 |
- |
- |
PREDICTED: glutathione transferase GST 23-like [Jatropha curcas] |
| 16 |
Hb_020332_040 |
0.126169439 |
- |
- |
PREDICTED: alcohol dehydrogenase 1-like [Jatropha curcas] |
| 17 |
Hb_001454_300 |
0.1339383353 |
- |
- |
nucleotide binding protein, putative [Ricinus communis] |
| 18 |
Hb_004195_240 |
0.1342598021 |
- |
- |
Peroxidase 72 precursor, putative [Ricinus communis] |
| 19 |
Hb_025469_040 |
0.134510469 |
- |
- |
PREDICTED: acidic mammalian chitinase-like [Jatropha curcas] |
| 20 |
Hb_001279_220 |
0.1360038507 |
- |
- |
protein binding protein, putative [Ricinus communis] |