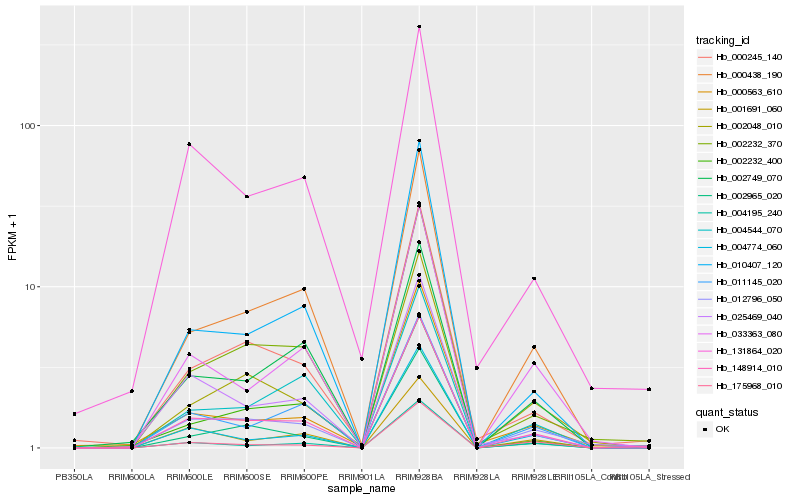
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_148914_010 |
0.0 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 2 |
Hb_012796_050 |
0.0636740525 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
1-deoxyxylulose-5-phosphate synthase, putative [Ricinus communis] |
| 3 |
Hb_000438_190 |
0.07922212 |
- |
- |
PREDICTED: peroxidase 4-like [Jatropha curcas] |
| 4 |
Hb_002232_370 |
0.0881725665 |
- |
- |
PREDICTED: receptor-like protein kinase HAIKU2 [Jatropha curcas] |
| 5 |
Hb_000245_140 |
0.0951501481 |
- |
- |
PREDICTED: CASP-like protein 3A1 [Jatropha curcas] |
| 6 |
Hb_002965_020 |
0.0954670584 |
- |
- |
hypothetical protein JCGZ_18947 [Jatropha curcas] |
| 7 |
Hb_010407_120 |
0.0987382844 |
- |
- |
PREDICTED: glutathione transferase GST 23-like [Jatropha curcas] |
| 8 |
Hb_004774_060 |
0.0987996907 |
- |
- |
- |
| 9 |
Hb_011145_020 |
0.100093776 |
- |
- |
PREDICTED: metalloendoproteinase 1-like [Pyrus x bretschneideri] |
| 10 |
Hb_025469_040 |
0.1031850268 |
- |
- |
PREDICTED: acidic mammalian chitinase-like [Jatropha curcas] |
| 11 |
Hb_033363_080 |
0.1071031467 |
- |
- |
Allene oxide cyclase 3, chloroplast precursor, putative [Ricinus communis] |
| 12 |
Hb_002048_010 |
0.1088376561 |
- |
- |
unknown [Lotus japonicus] |
| 13 |
Hb_000563_610 |
0.1164118439 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase LIN-1 [Jatropha curcas] |
| 14 |
Hb_002749_070 |
0.1206261012 |
- |
- |
PREDICTED: uncharacterized protein LOC105635941 [Jatropha curcas] |
| 15 |
Hb_001691_060 |
0.1220513589 |
- |
- |
lipoxygenase, putative [Ricinus communis] |
| 16 |
Hb_175968_010 |
0.1234725333 |
- |
- |
hypothetical protein JCGZ_21281 [Jatropha curcas] |
| 17 |
Hb_004544_070 |
0.1245611751 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 18 |
Hb_004195_240 |
0.1248204999 |
- |
- |
Peroxidase 72 precursor, putative [Ricinus communis] |
| 19 |
Hb_002232_400 |
0.1248961108 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH25-like [Jatropha curcas] |
| 20 |
Hb_131864_020 |
0.125229684 |
- |
- |
PREDICTED: anthocyanidin reductase [Jatropha curcas] |