| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
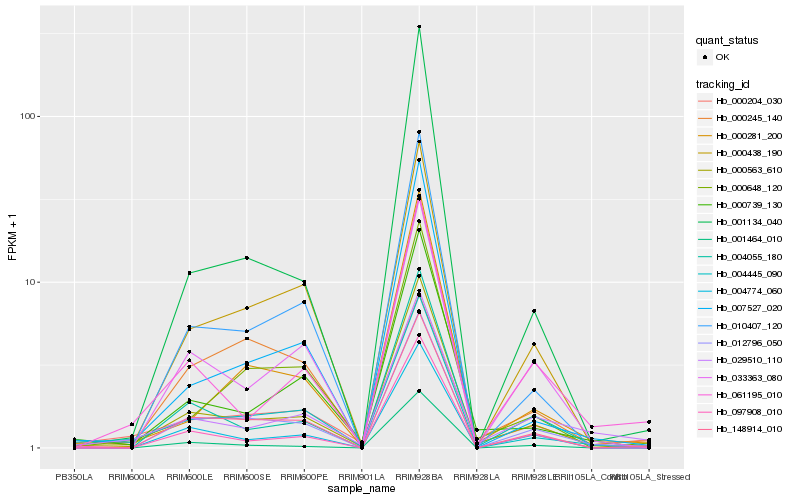
Hb_000563_610 |
0.0 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase LIN-1 [Jatropha curcas] |
| 2 |
Hb_004055_180 |
0.0923736146 |
- |
- |
cell wall invertase [Manihot esculenta] |
| 3 |
Hb_000739_130 |
0.0983637337 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000245_140 |
0.1009922203 |
- |
- |
PREDICTED: CASP-like protein 3A1 [Jatropha curcas] |
| 5 |
Hb_097908_010 |
0.1049688764 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g03230 isoform X2 [Jatropha curcas] |
| 6 |
Hb_012796_050 |
0.1116004794 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
1-deoxyxylulose-5-phosphate synthase, putative [Ricinus communis] |
| 7 |
Hb_004774_060 |
0.1130260666 |
- |
- |
- |
| 8 |
Hb_004445_090 |
0.1144375226 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000204_030 |
0.1151619948 |
- |
- |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 10 |
Hb_148914_010 |
0.1164118439 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 11 |
Hb_010407_120 |
0.1178595374 |
- |
- |
PREDICTED: glutathione transferase GST 23-like [Jatropha curcas] |
| 12 |
Hb_029510_110 |
0.1212770974 |
- |
- |
PREDICTED: protein PLANT CADMIUM RESISTANCE 8-like [Jatropha curcas] |
| 13 |
Hb_000281_200 |
0.1213865368 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 [Jatropha curcas] |
| 14 |
Hb_061195_010 |
0.1250658072 |
- |
- |
Galactose oxidase precursor, putative [Ricinus communis] |
| 15 |
Hb_007527_020 |
0.1254992693 |
- |
- |
PREDICTED: feruloyl CoA ortho-hydroxylase 1 [Jatropha curcas] |
| 16 |
Hb_000648_120 |
0.1255459798 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase IV.1 [Vitis vinifera] |
| 17 |
Hb_033363_080 |
0.1262155406 |
- |
- |
Allene oxide cyclase 3, chloroplast precursor, putative [Ricinus communis] |
| 18 |
Hb_001464_010 |
0.1267147407 |
- |
- |
PREDICTED: putative receptor-like protein kinase At4g00960 isoform X1 [Populus euphratica] |
| 19 |
Hb_001134_040 |
0.1271897694 |
- |
- |
PREDICTED: EG45-like domain containing protein [Jatropha curcas] |
| 20 |
Hb_000438_190 |
0.1287333134 |
- |
- |
PREDICTED: peroxidase 4-like [Jatropha curcas] |