| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
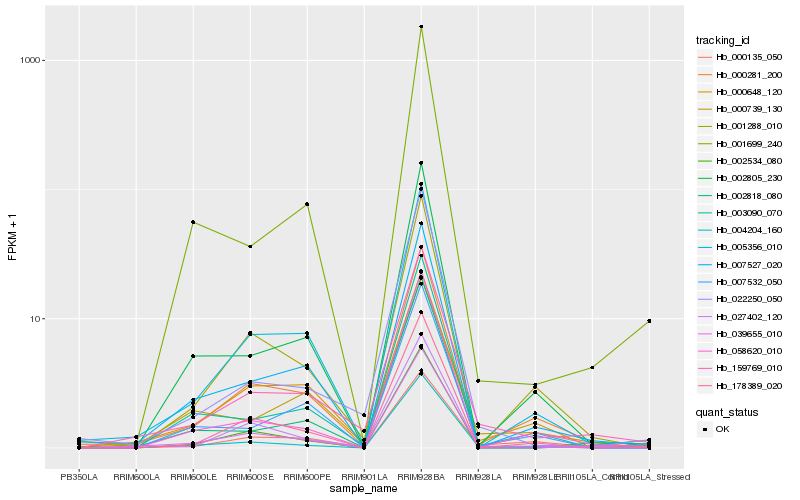
Hb_159769_010 |
0.0 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 2 |
Hb_039655_010 |
0.1102106016 |
- |
- |
PREDICTED: BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1-like [Malus domestica] |
| 3 |
Hb_000281_200 |
0.1108604533 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 [Jatropha curcas] |
| 4 |
Hb_000135_050 |
0.1195034609 |
- |
- |
PREDICTED: putative leucine-rich repeat receptor-like protein kinase At2g19210-like isoform X2 [Citrus sinensis] |
| 5 |
Hb_022250_050 |
0.125547345 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_005356_010 |
0.1268043275 |
- |
- |
hypothetical protein POPTR_0003s16550g [Populus trichocarpa] |
| 7 |
Hb_007527_020 |
0.127279144 |
- |
- |
PREDICTED: feruloyl CoA ortho-hydroxylase 1 [Jatropha curcas] |
| 8 |
Hb_004204_160 |
0.1318181089 |
- |
- |
S-locus-specific glycoprotein S6 precursor, putative [Ricinus communis] |
| 9 |
Hb_000739_130 |
0.1334672624 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000648_120 |
0.1335275257 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase IV.1 [Vitis vinifera] |
| 11 |
Hb_001288_010 |
0.139434966 |
- |
- |
- |
| 12 |
Hb_001699_240 |
0.1396032396 |
- |
- |
PREDICTED: naringenin,2-oxoglutarate 3-dioxygenase [Jatropha curcas] |
| 13 |
Hb_027402_120 |
0.1398577371 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 14 |
Hb_002805_230 |
0.1413340437 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12 isoform X1 [Jatropha curcas] |
| 15 |
Hb_003090_070 |
0.1423071689 |
- |
- |
PREDICTED: laccase-7 [Jatropha curcas] |
| 16 |
Hb_002534_080 |
0.1428606719 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 17 |
Hb_178389_020 |
0.1436209102 |
- |
- |
PREDICTED: cytochrome P450 86B1-like [Jatropha curcas] |
| 18 |
Hb_002818_080 |
0.1439321048 |
transcription factor |
TF Family: AUX/IAA |
Auxin-responsive protein IAA20, putative [Ricinus communis] |
| 19 |
Hb_058620_010 |
0.1466694959 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 20 |
Hb_007532_050 |
0.1478384265 |
- |
- |
9-cis-epoxycarotenoid dioxygenase, putative [Ricinus communis] |