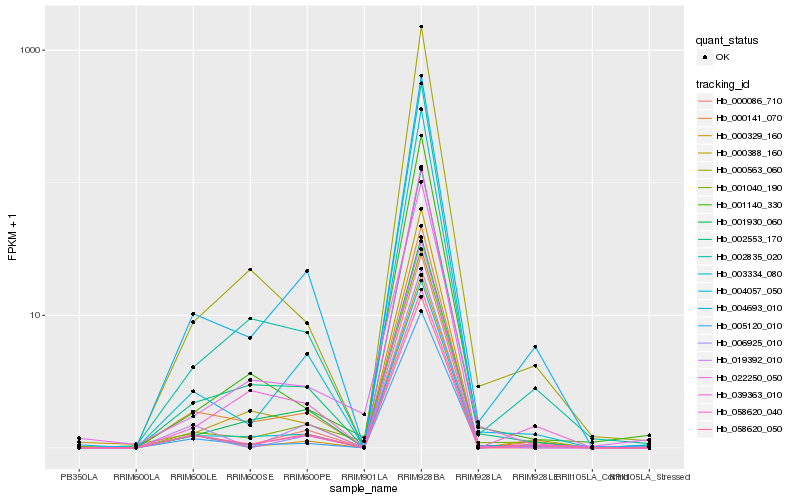
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001040_190 |
0.0 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor B-2a-like [Jatropha curcas] |
| 2 |
Hb_058620_040 |
0.0676807524 |
- |
- |
Myrcene synthase, chloroplastic, putative isoform 1 [Theobroma cacao] |
| 3 |
Hb_002553_170 |
0.0692331535 |
- |
- |
PREDICTED: early nodulin-75-like [Jatropha curcas] |
| 4 |
Hb_003334_080 |
0.0696156712 |
- |
- |
Reticuline oxidase precursor, putative [Ricinus communis] |
| 5 |
Hb_058620_050 |
0.0717581152 |
- |
- |
PREDICTED: terpene synthase 10-like [Jatropha curcas] |
| 6 |
Hb_000141_070 |
0.0718794448 |
- |
- |
l-lactate dehydrogenase, putative [Ricinus communis] |
| 7 |
Hb_004693_010 |
0.0731783839 |
- |
- |
hypothetical protein JCGZ_02321 [Jatropha curcas] |
| 8 |
Hb_002835_020 |
0.0744572334 |
- |
- |
PREDICTED: cytochrome P450 71A1-like [Jatropha curcas] |
| 9 |
Hb_000329_160 |
0.0746359893 |
- |
- |
PREDICTED: pectinesterase 2 isoform X1 [Jatropha curcas] |
| 10 |
Hb_004057_050 |
0.0747170646 |
- |
- |
PREDICTED: cytochrome P450 76A1-like [Jatropha curcas] |
| 11 |
Hb_005120_010 |
0.0773685693 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 12 |
Hb_001930_060 |
0.0787718545 |
- |
- |
PREDICTED: cytochrome P450 71D9-like [Fragaria vesca subsp. vesca] |
| 13 |
Hb_022250_050 |
0.0792471481 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000086_710 |
0.0799709164 |
- |
- |
PREDICTED: feruloyl CoA ortho-hydroxylase 2-like [Jatropha curcas] |
| 15 |
Hb_000563_060 |
0.081831014 |
- |
- |
PREDICTED: uncharacterized protein LOC100306381 isoform X1 [Glycine max] |
| 16 |
Hb_039363_010 |
0.0831848795 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 17 |
Hb_000388_160 |
0.0833823058 |
- |
- |
xyloglucan endo-1 family protein [Populus trichocarpa] |
| 18 |
Hb_001140_330 |
0.0839558554 |
- |
- |
PREDICTED: uncharacterized protein LOC105142675 [Populus euphratica] |
| 19 |
Hb_019392_010 |
0.0845803416 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable S-adenosylmethionine-dependent methyltransferase At5g38100 [Jatropha curcas] |
| 20 |
Hb_006925_010 |
0.0858872572 |
- |
- |
PREDICTED: uncharacterized protein LOC101777526 [Setaria italica] |