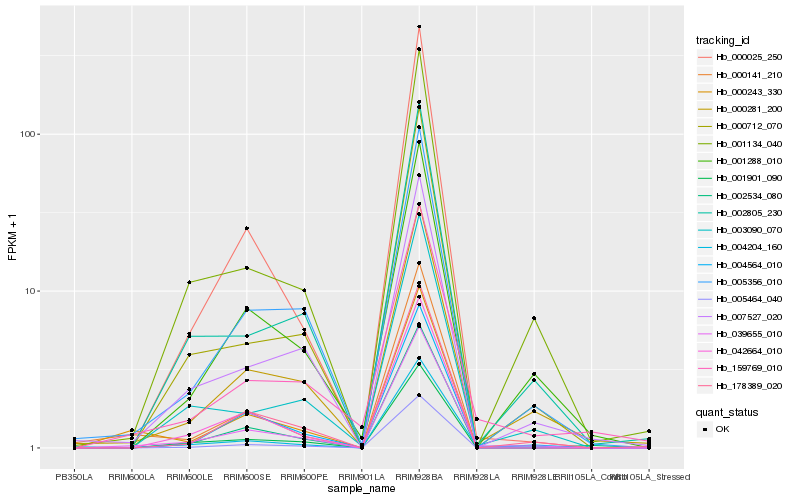
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_039655_010 |
0.0 |
- |
- |
PREDICTED: BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1-like [Malus domestica] |
| 2 |
Hb_004204_160 |
0.0768917668 |
- |
- |
S-locus-specific glycoprotein S6 precursor, putative [Ricinus communis] |
| 3 |
Hb_005356_010 |
0.0901895051 |
- |
- |
hypothetical protein POPTR_0003s16550g [Populus trichocarpa] |
| 4 |
Hb_000281_200 |
0.0915049631 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 [Jatropha curcas] |
| 5 |
Hb_178389_020 |
0.0936159871 |
- |
- |
PREDICTED: cytochrome P450 86B1-like [Jatropha curcas] |
| 6 |
Hb_004564_010 |
0.0966404848 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 7 |
Hb_001134_040 |
0.0979839132 |
- |
- |
PREDICTED: EG45-like domain containing protein [Jatropha curcas] |
| 8 |
Hb_001901_090 |
0.1005498689 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 9 |
Hb_042664_010 |
0.1010212479 |
- |
- |
hypothetical protein POPTR_0001s07640g, partial [Populus trichocarpa] |
| 10 |
Hb_005464_040 |
0.1020025323 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 11 |
Hb_000712_070 |
0.1031453652 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001288_010 |
0.1053262239 |
- |
- |
- |
| 13 |
Hb_007527_020 |
0.1057649028 |
- |
- |
PREDICTED: feruloyl CoA ortho-hydroxylase 1 [Jatropha curcas] |
| 14 |
Hb_002805_230 |
0.1068628177 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000243_330 |
0.1069496806 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000025_250 |
0.1086311758 |
- |
- |
PREDICTED: flavonoid 3',5'-hydroxylase 2-like [Jatropha curcas] |
| 17 |
Hb_003090_070 |
0.1093111646 |
- |
- |
PREDICTED: laccase-7 [Jatropha curcas] |
| 18 |
Hb_002534_080 |
0.109466998 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 19 |
Hb_159769_010 |
0.1102106016 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 20 |
Hb_000141_210 |
0.1114718693 |
- |
- |
PREDICTED: 36.4 kDa proline-rich protein-like [Camelina sativa] |