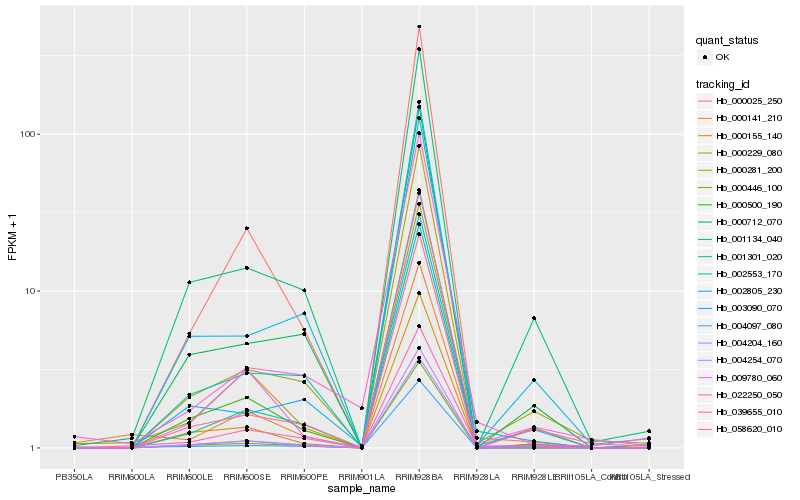
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004204_160 |
0.0 |
- |
- |
S-locus-specific glycoprotein S6 precursor, putative [Ricinus communis] |
| 2 |
Hb_039655_010 |
0.0768917668 |
- |
- |
PREDICTED: BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1-like [Malus domestica] |
| 3 |
Hb_001134_040 |
0.0960041823 |
- |
- |
PREDICTED: EG45-like domain containing protein [Jatropha curcas] |
| 4 |
Hb_004097_080 |
0.0984450647 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 isoform X1 [Nicotiana tomentosiformis] |
| 5 |
Hb_000712_070 |
0.1018406046 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_058620_010 |
0.1020776765 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 7 |
Hb_003090_070 |
0.1042974389 |
- |
- |
PREDICTED: laccase-7 [Jatropha curcas] |
| 8 |
Hb_009780_060 |
0.1047781117 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranyl geranyl pyrophosphate synthase, putative [Ricinus communis] |
| 9 |
Hb_002805_230 |
0.1078884041 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000281_200 |
0.1081256145 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 [Jatropha curcas] |
| 11 |
Hb_000155_140 |
0.1103497618 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 12 |
Hb_000229_080 |
0.1112780427 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 13 |
Hb_002553_170 |
0.11197859 |
- |
- |
PREDICTED: early nodulin-75-like [Jatropha curcas] |
| 14 |
Hb_022250_050 |
0.1123259949 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000025_250 |
0.1132221309 |
- |
- |
PREDICTED: flavonoid 3',5'-hydroxylase 2-like [Jatropha curcas] |
| 16 |
Hb_000500_190 |
0.1138790201 |
- |
- |
hypothetical protein JCGZ_21655 [Jatropha curcas] |
| 17 |
Hb_000446_100 |
0.1150017129 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4-like [Jatropha curcas] |
| 18 |
Hb_004254_070 |
0.1157630832 |
transcription factor |
TF Family: NAC |
hypothetical protein POPTR_0015s03700g [Populus trichocarpa] |
| 19 |
Hb_000141_210 |
0.1159650559 |
- |
- |
PREDICTED: 36.4 kDa proline-rich protein-like [Camelina sativa] |
| 20 |
Hb_001301_020 |
0.1170156117 |
- |
- |
hypothetical protein JCGZ_06575 [Jatropha curcas] |