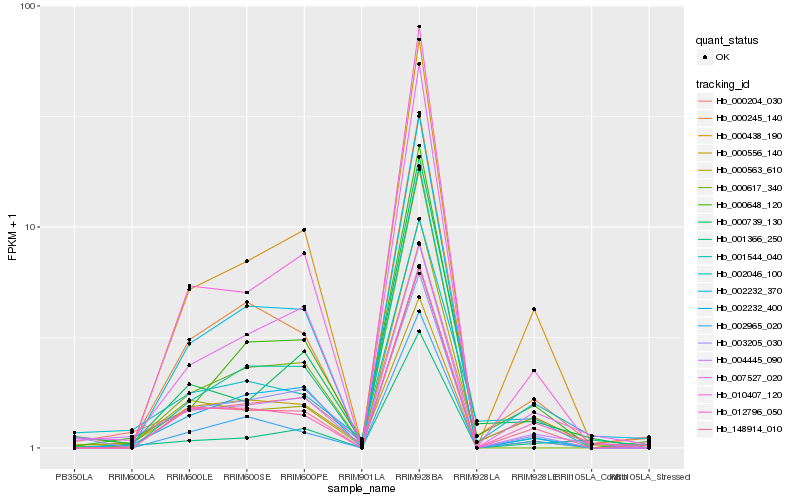
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004445_090 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000245_140 |
0.1117618445 |
- |
- |
PREDICTED: CASP-like protein 3A1 [Jatropha curcas] |
| 3 |
Hb_000563_610 |
0.1144375226 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase LIN-1 [Jatropha curcas] |
| 4 |
Hb_000204_030 |
0.1187639255 |
- |
- |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 5 |
Hb_002232_370 |
0.1241106371 |
- |
- |
PREDICTED: receptor-like protein kinase HAIKU2 [Jatropha curcas] |
| 6 |
Hb_000739_130 |
0.1262768775 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000438_190 |
0.1272738246 |
- |
- |
PREDICTED: peroxidase 4-like [Jatropha curcas] |
| 8 |
Hb_010407_120 |
0.1286416154 |
- |
- |
PREDICTED: glutathione transferase GST 23-like [Jatropha curcas] |
| 9 |
Hb_002232_400 |
0.129389081 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH25-like [Jatropha curcas] |
| 10 |
Hb_001366_250 |
0.1314733456 |
- |
- |
PREDICTED: mannan endo-1,4-beta-mannosidase 4-like [Jatropha curcas] |
| 11 |
Hb_000648_120 |
0.134067561 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase IV.1 [Vitis vinifera] |
| 12 |
Hb_007527_020 |
0.1343543685 |
- |
- |
PREDICTED: feruloyl CoA ortho-hydroxylase 1 [Jatropha curcas] |
| 13 |
Hb_003205_030 |
0.1355291631 |
- |
- |
Pectinesterase inhibitor, putative [Ricinus communis] |
| 14 |
Hb_001544_040 |
0.1384975585 |
- |
- |
PREDICTED: uncharacterized protein At1g66480-like [Populus euphratica] |
| 15 |
Hb_012796_050 |
0.1402499322 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
1-deoxyxylulose-5-phosphate synthase, putative [Ricinus communis] |
| 16 |
Hb_148914_010 |
0.1406243326 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 17 |
Hb_000556_140 |
0.1408769251 |
transcription factor |
TF Family: GRF |
PREDICTED: growth-regulating factor 9 [Jatropha curcas] |
| 18 |
Hb_000617_340 |
0.1417050458 |
- |
- |
respiratory burst oxidase B [Manihot esculenta] |
| 19 |
Hb_002046_100 |
0.1421684089 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB82 [Jatropha curcas] |
| 20 |
Hb_002965_020 |
0.1444109764 |
- |
- |
hypothetical protein JCGZ_18947 [Jatropha curcas] |