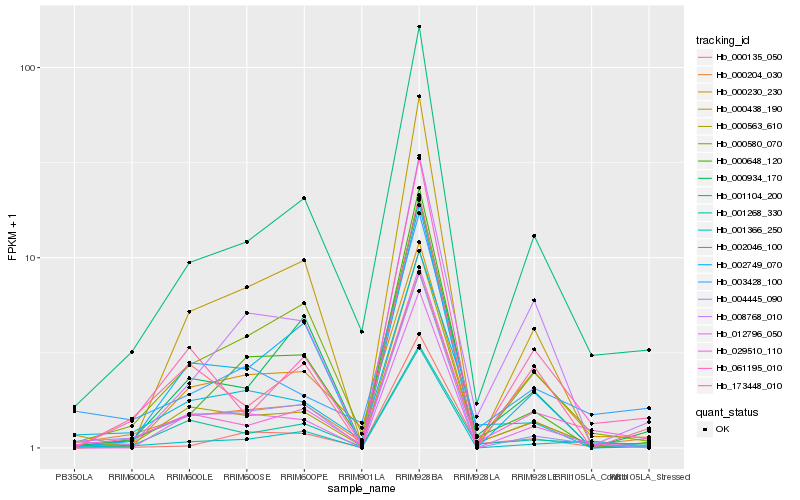
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001104_200 |
0.0 |
- |
- |
Phytosulfokine 3 precursor, putative [Theobroma cacao] |
| 2 |
Hb_000204_030 |
0.0766790945 |
- |
- |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 3 |
Hb_029510_110 |
0.0985956472 |
- |
- |
PREDICTED: protein PLANT CADMIUM RESISTANCE 8-like [Jatropha curcas] |
| 4 |
Hb_000230_230 |
0.124231091 |
- |
- |
hydrolase, putative [Ricinus communis] |
| 5 |
Hb_000934_170 |
0.1326208159 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 6 |
Hb_000563_610 |
0.1375361021 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase LIN-1 [Jatropha curcas] |
| 7 |
Hb_000580_070 |
0.1399393351 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g10290 [Jatropha curcas] |
| 8 |
Hb_004445_090 |
0.1464726482 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_061195_010 |
0.1486435038 |
- |
- |
Galactose oxidase precursor, putative [Ricinus communis] |
| 10 |
Hb_000648_120 |
0.1489326055 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase IV.1 [Vitis vinifera] |
| 11 |
Hb_173448_010 |
0.1499440751 |
- |
- |
PREDICTED: galactose oxidase-like [Jatropha curcas] |
| 12 |
Hb_001268_330 |
0.1518829396 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 13 |
Hb_001366_250 |
0.152056533 |
- |
- |
PREDICTED: mannan endo-1,4-beta-mannosidase 4-like [Jatropha curcas] |
| 14 |
Hb_000438_190 |
0.152456897 |
- |
- |
PREDICTED: peroxidase 4-like [Jatropha curcas] |
| 15 |
Hb_003428_100 |
0.1528339711 |
- |
- |
Polygalacturonase precursor, putative [Ricinus communis] |
| 16 |
Hb_008768_010 |
0.1538868901 |
- |
- |
PREDICTED: equilibrative nucleotide transporter 8 isoform X1 [Jatropha curcas] |
| 17 |
Hb_012796_050 |
0.155720298 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
1-deoxyxylulose-5-phosphate synthase, putative [Ricinus communis] |
| 18 |
Hb_002046_100 |
0.1597128264 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB82 [Jatropha curcas] |
| 19 |
Hb_000135_050 |
0.1622036764 |
- |
- |
PREDICTED: putative leucine-rich repeat receptor-like protein kinase At2g19210-like isoform X2 [Citrus sinensis] |
| 20 |
Hb_002749_070 |
0.1630376415 |
- |
- |
PREDICTED: uncharacterized protein LOC105635941 [Jatropha curcas] |