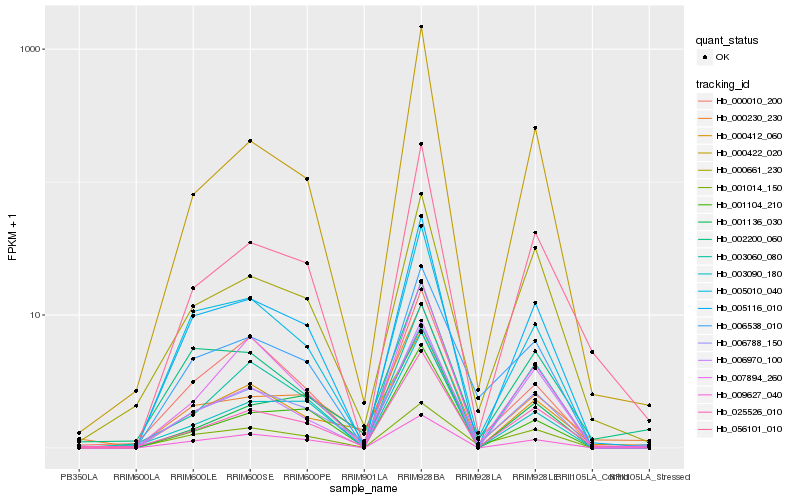
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006788_150 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_025526_010 |
0.0836950897 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104452086 [Eucalyptus grandis] |
| 3 |
Hb_005116_010 |
0.1133783531 |
transcription factor |
TF Family: bHLH |
PREDICTED: basic helix-loop-helix protein A [Jatropha curcas] |
| 4 |
Hb_000661_230 |
0.1149306 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 22-like isoform X2 [Jatropha curcas] |
| 5 |
Hb_000412_060 |
0.1172573684 |
- |
- |
hypothetical protein JCGZ_18814 [Jatropha curcas] |
| 6 |
Hb_007894_260 |
0.1201437898 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 7 |
Hb_005010_040 |
0.1234803608 |
- |
- |
PREDICTED: uncharacterized protein LOC105630824 [Jatropha curcas] |
| 8 |
Hb_006538_010 |
0.1247361826 |
- |
- |
PREDICTED: universal stress protein A-like protein [Jatropha curcas] |
| 9 |
Hb_001014_150 |
0.1273625678 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 10 |
Hb_000010_200 |
0.1290925653 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 11 |
Hb_003060_080 |
0.1296286225 |
- |
- |
PREDICTED: acyl-CoA--sterol O-acyltransferase 1 [Jatropha curcas] |
| 12 |
Hb_009627_040 |
0.1309069465 |
- |
- |
Lipase class 3-related protein, putative [Theobroma cacao] |
| 13 |
Hb_000230_230 |
0.1318478942 |
- |
- |
hydrolase, putative [Ricinus communis] |
| 14 |
Hb_003090_180 |
0.1321231317 |
- |
- |
PREDICTED: uncharacterized protein LOC105646674 [Jatropha curcas] |
| 15 |
Hb_056101_010 |
0.1343889427 |
- |
- |
hypothetical protein POPTR_0391s00200g [Populus trichocarpa] |
| 16 |
Hb_002200_060 |
0.1385797892 |
- |
- |
PREDICTED: pleckstrin homology domain-containing family A member 8 [Jatropha curcas] |
| 17 |
Hb_000422_020 |
0.1387472472 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 18 |
Hb_001104_210 |
0.1404670212 |
- |
- |
hypothetical protein JCGZ_23620 [Jatropha curcas] |
| 19 |
Hb_006970_100 |
0.1411764752 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 20 |
Hb_001136_030 |
0.1417666606 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 25-like [Jatropha curcas] |