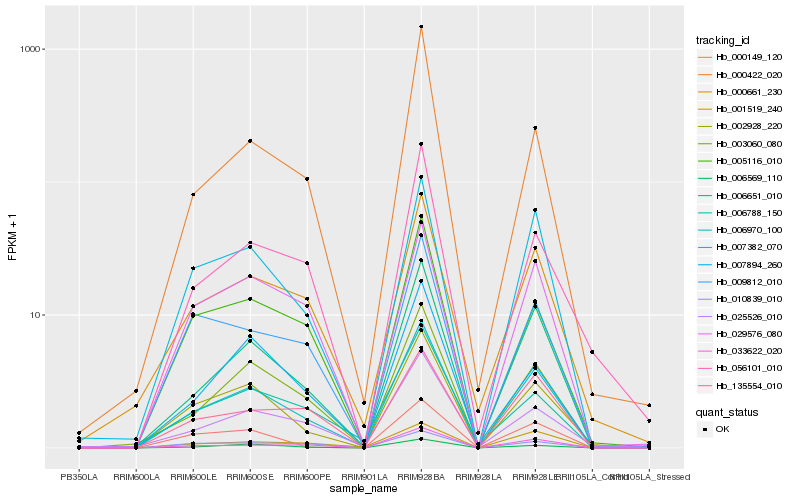
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006970_100 |
0.0 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 2 |
Hb_006651_010 |
0.0682560287 |
- |
- |
PREDICTED: protein disulfide isomerase-like 1-4-like [Citrus sinensis] |
| 3 |
Hb_007382_070 |
0.0937856516 |
- |
- |
PREDICTED: MAPK-interacting and spindle-stabilizing protein-like [Jatropha curcas] |
| 4 |
Hb_002928_220 |
0.0974299298 |
- |
- |
serine/threonine-protein kinase cx32, putative [Ricinus communis] |
| 5 |
Hb_003060_080 |
0.0981889267 |
- |
- |
PREDICTED: acyl-CoA--sterol O-acyltransferase 1 [Jatropha curcas] |
| 6 |
Hb_033622_020 |
0.1068361397 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 7 |
Hb_006569_110 |
0.116706767 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000661_230 |
0.1181141838 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 22-like isoform X2 [Jatropha curcas] |
| 9 |
Hb_005116_010 |
0.1238952678 |
transcription factor |
TF Family: bHLH |
PREDICTED: basic helix-loop-helix protein A [Jatropha curcas] |
| 10 |
Hb_009812_010 |
0.1242017585 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 11 |
Hb_135554_010 |
0.125208782 |
- |
- |
Insulin-degrading enzyme, putative [Ricinus communis] |
| 12 |
Hb_025526_010 |
0.1304652038 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104452086 [Eucalyptus grandis] |
| 13 |
Hb_001519_240 |
0.1305213008 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 14 |
Hb_029576_080 |
0.132421125 |
- |
- |
Aspartic proteinase precursor, putative [Ricinus communis] |
| 15 |
Hb_000149_120 |
0.1369282224 |
- |
- |
PREDICTED: uncharacterized protein LOC105642119 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000422_020 |
0.1383650458 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 17 |
Hb_006788_150 |
0.1411764752 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_010839_010 |
0.1415546632 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 19 |
Hb_007894_260 |
0.141946201 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 20 |
Hb_056101_010 |
0.142160688 |
- |
- |
hypothetical protein POPTR_0391s00200g [Populus trichocarpa] |