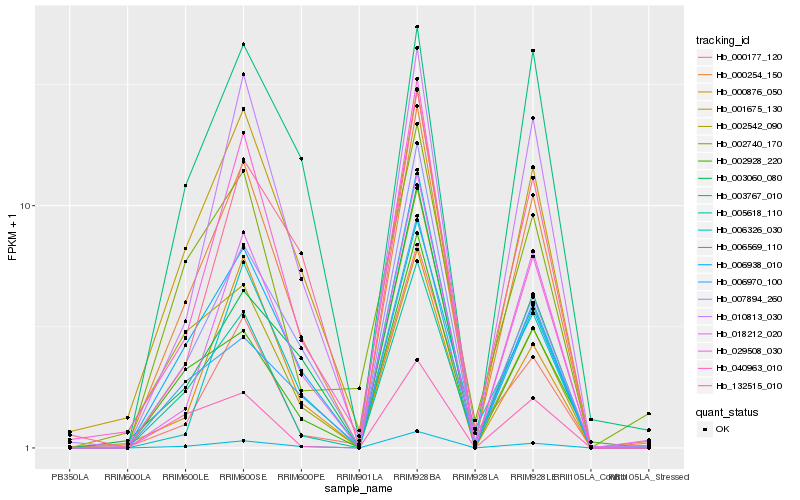
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000254_150 |
0.0 |
- |
- |
PREDICTED: homeobox-leucine zipper protein GLABRA 2 [Jatropha curcas] |
| 2 |
Hb_006569_110 |
0.0848247236 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_018212_020 |
0.0967963894 |
- |
- |
PREDICTED: uncharacterized protein LOC105636321 [Jatropha curcas] |
| 4 |
Hb_010813_030 |
0.0981400292 |
- |
- |
PREDICTED: indole-3-acetaldehyde oxidase-like [Jatropha curcas] |
| 5 |
Hb_005618_110 |
0.1026759673 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 6 |
Hb_001675_130 |
0.1029295087 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein GLABRA 2 [Jatropha curcas] |
| 7 |
Hb_006938_010 |
0.1080265495 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB3-like [Jatropha curcas] |
| 8 |
Hb_132515_010 |
0.1321062182 |
- |
- |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 9 |
Hb_003060_080 |
0.1324279997 |
- |
- |
PREDICTED: acyl-CoA--sterol O-acyltransferase 1 [Jatropha curcas] |
| 10 |
Hb_002928_220 |
0.1339164165 |
- |
- |
serine/threonine-protein kinase cx32, putative [Ricinus communis] |
| 11 |
Hb_007894_260 |
0.136657258 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 12 |
Hb_000876_050 |
0.1409487709 |
- |
- |
PREDICTED: heparan-alpha-glucosaminide N-acetyltransferase-like [Jatropha curcas] |
| 13 |
Hb_002740_170 |
0.1418199637 |
- |
- |
PREDICTED: uncharacterized protein LOC105647357 [Jatropha curcas] |
| 14 |
Hb_006326_030 |
0.1439574045 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_040963_010 |
0.148908312 |
- |
- |
PREDICTED: polyneuridine-aldehyde esterase-like [Populus euphratica] |
| 16 |
Hb_006970_100 |
0.1518576934 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 17 |
Hb_029508_030 |
0.1529543495 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0002s04880g [Populus trichocarpa] |
| 18 |
Hb_003767_010 |
0.1531257317 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD11 [Jatropha curcas] |
| 19 |
Hb_000177_120 |
0.1550125614 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 isoform X1 [Jatropha curcas] |
| 20 |
Hb_002542_090 |
0.1555246565 |
- |
- |
Potassium transporter, putative [Ricinus communis] |