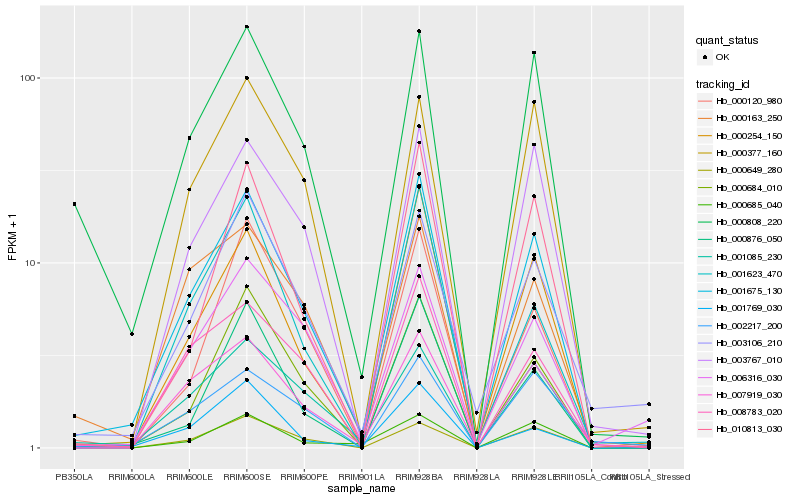
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001675_130 |
0.0 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein GLABRA 2 [Jatropha curcas] |
| 2 |
Hb_000254_150 |
0.1029295087 |
- |
- |
PREDICTED: homeobox-leucine zipper protein GLABRA 2 [Jatropha curcas] |
| 3 |
Hb_007919_030 |
0.1041126233 |
- |
- |
- |
| 4 |
Hb_000684_010 |
0.1177250344 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 5 |
Hb_003767_010 |
0.1259745937 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD11 [Jatropha curcas] |
| 6 |
Hb_000649_280 |
0.1285987024 |
- |
- |
PREDICTED: F-box protein PP2-B10-like [Pyrus x bretschneideri] |
| 7 |
Hb_010813_030 |
0.1289357905 |
- |
- |
PREDICTED: indole-3-acetaldehyde oxidase-like [Jatropha curcas] |
| 8 |
Hb_001085_230 |
0.1291263809 |
- |
- |
PREDICTED: receptor like protein kinase S.2 [Jatropha curcas] |
| 9 |
Hb_000876_050 |
0.1356353073 |
- |
- |
PREDICTED: heparan-alpha-glucosaminide N-acetyltransferase-like [Jatropha curcas] |
| 10 |
Hb_000808_220 |
0.1359707002 |
- |
- |
PREDICTED: pyrophosphate-energized vacuolar membrane proton pump 1-like [Jatropha curcas] |
| 11 |
Hb_000377_160 |
0.1370440708 |
- |
- |
PREDICTED: oligopeptide transporter 3 [Jatropha curcas] |
| 12 |
Hb_001623_470 |
0.1409131051 |
- |
- |
ammonium transporter, putative [Ricinus communis] |
| 13 |
Hb_008783_020 |
0.1418982659 |
- |
- |
hypothetical protein JCGZ_21284 [Jatropha curcas] |
| 14 |
Hb_000163_250 |
0.1439787288 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 15 |
Hb_000120_980 |
0.1446277368 |
- |
- |
PREDICTED: protein STRICTOSIDINE SYNTHASE-LIKE 5-like [Jatropha curcas] |
| 16 |
Hb_006316_030 |
0.1447766153 |
- |
- |
aldehyde oxidase, putative [Ricinus communis] |
| 17 |
Hb_002217_200 |
0.1451091312 |
- |
- |
PREDICTED: gibberellin 2-beta-dioxygenase 2 [Jatropha curcas] |
| 18 |
Hb_001769_030 |
0.1456204593 |
- |
- |
PREDICTED: exocyst complex component EXO70A1-like [Jatropha curcas] |
| 19 |
Hb_003106_210 |
0.1458187836 |
- |
- |
PREDICTED: UDP-glycosyltransferase 74B1-like [Jatropha curcas] |
| 20 |
Hb_000685_040 |
0.1522050793 |
- |
- |
hypothetical protein OsJ_17676 [Oryza sativa Japonica Group] |