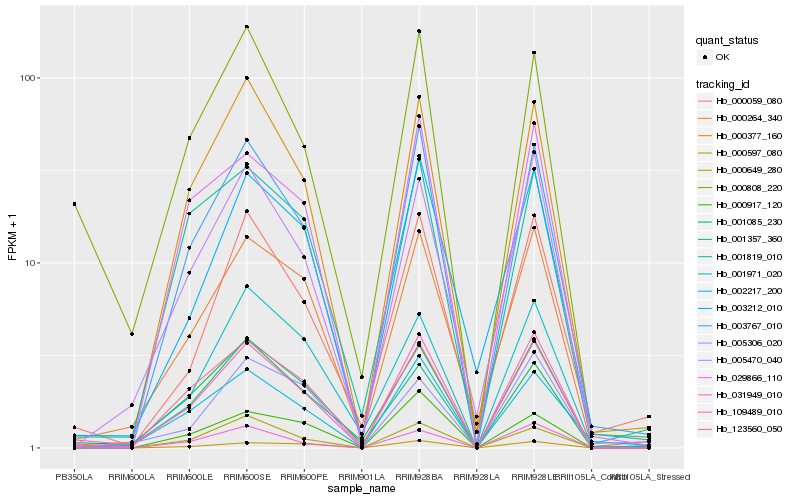
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_109489_010 |
0.0 |
- |
- |
multidrug resistance-associated protein 2, 6 (mrp2, 6), abc-transoprter, putative [Ricinus communis] |
| 2 |
Hb_003767_010 |
0.1026227397 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD11 [Jatropha curcas] |
| 3 |
Hb_005470_040 |
0.1060199623 |
- |
- |
hypothetical protein POPTR_0015s09760g [Populus trichocarpa] |
| 4 |
Hb_000264_340 |
0.1076995283 |
- |
- |
PREDICTED: callose synthase 11-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_123560_050 |
0.109917725 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g24010 [Jatropha curcas] |
| 6 |
Hb_029866_110 |
0.1185488397 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: ABC transporter C family member 10-like [Jatropha curcas] |
| 7 |
Hb_000808_220 |
0.118895674 |
- |
- |
PREDICTED: pyrophosphate-energized vacuolar membrane proton pump 1-like [Jatropha curcas] |
| 8 |
Hb_000377_160 |
0.1199150964 |
- |
- |
PREDICTED: oligopeptide transporter 3 [Jatropha curcas] |
| 9 |
Hb_002217_200 |
0.1228350553 |
- |
- |
PREDICTED: gibberellin 2-beta-dioxygenase 2 [Jatropha curcas] |
| 10 |
Hb_031949_010 |
0.1232536699 |
- |
- |
- |
| 11 |
Hb_000597_080 |
0.1254536457 |
- |
- |
PREDICTED: 7-deoxyloganetic acid glucosyltransferase-like isoform X2 [Jatropha curcas] |
| 12 |
Hb_003212_010 |
0.1289538383 |
- |
- |
sucrose transporter 4 [Hevea brasiliensis] |
| 13 |
Hb_001085_230 |
0.1342983834 |
- |
- |
PREDICTED: receptor like protein kinase S.2 [Jatropha curcas] |
| 14 |
Hb_000059_080 |
0.1355669419 |
- |
- |
PREDICTED: uncharacterized protein LOC105629177 [Jatropha curcas] |
| 15 |
Hb_000649_280 |
0.1409737911 |
- |
- |
PREDICTED: F-box protein PP2-B10-like [Pyrus x bretschneideri] |
| 16 |
Hb_005306_020 |
0.1418374398 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB86 [Jatropha curcas] |
| 17 |
Hb_001819_010 |
0.1427737556 |
- |
- |
PREDICTED: potassium transporter 8 [Jatropha curcas] |
| 18 |
Hb_001357_360 |
0.1439002938 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 19 |
Hb_001971_020 |
0.1506138756 |
- |
- |
PREDICTED: alpha-dioxygenase 2 [Populus euphratica] |
| 20 |
Hb_000917_120 |
0.1516078225 |
- |
- |
conserved hypothetical protein [Ricinus communis] |