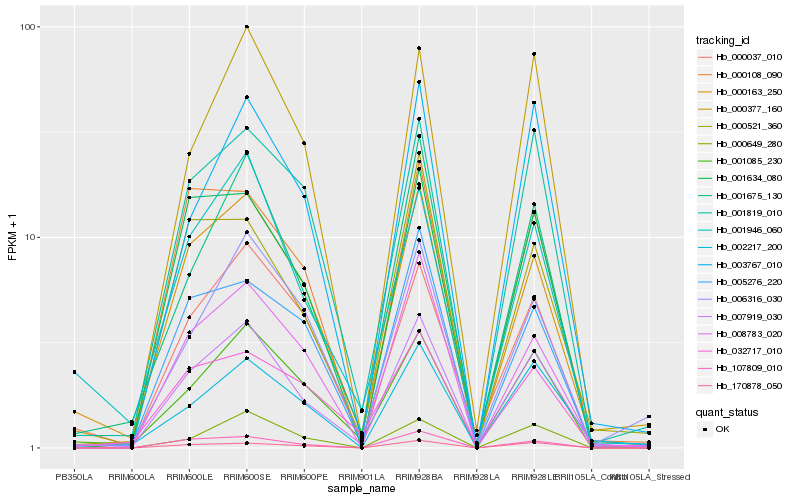
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007919_030 |
0.0 |
- |
- |
- |
| 2 |
Hb_000163_250 |
0.0862271405 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 3 |
Hb_001675_130 |
0.1041126233 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein GLABRA 2 [Jatropha curcas] |
| 4 |
Hb_001085_230 |
0.1081012491 |
- |
- |
PREDICTED: receptor like protein kinase S.2 [Jatropha curcas] |
| 5 |
Hb_001946_060 |
0.1198158705 |
- |
- |
PREDICTED: TBC1 domain family member 13-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_002217_200 |
0.120390992 |
- |
- |
PREDICTED: gibberellin 2-beta-dioxygenase 2 [Jatropha curcas] |
| 7 |
Hb_001819_010 |
0.1216149429 |
- |
- |
PREDICTED: potassium transporter 8 [Jatropha curcas] |
| 8 |
Hb_008783_020 |
0.1232358961 |
- |
- |
hypothetical protein JCGZ_21284 [Jatropha curcas] |
| 9 |
Hb_107809_010 |
0.1252825662 |
- |
- |
JHL06P13.14 [Jatropha curcas] |
| 10 |
Hb_000377_160 |
0.125806164 |
- |
- |
PREDICTED: oligopeptide transporter 3 [Jatropha curcas] |
| 11 |
Hb_000037_010 |
0.1278863749 |
- |
- |
PREDICTED: cation/H(+) antiporter 18-like [Jatropha curcas] |
| 12 |
Hb_003767_010 |
0.132130848 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD11 [Jatropha curcas] |
| 13 |
Hb_032717_010 |
0.1322036855 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 14 |
Hb_001634_080 |
0.1333985554 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 15 |
Hb_170878_050 |
0.1334826424 |
- |
- |
retrotransposon protein, putative, unclassified [Oryza sativa Japonica Group] |
| 16 |
Hb_006316_030 |
0.1360930164 |
- |
- |
aldehyde oxidase, putative [Ricinus communis] |
| 17 |
Hb_005276_220 |
0.1366796247 |
transcription factor |
TF Family: bHLH |
PREDICTED: basic helix-loop-helix protein A [Jatropha curcas] |
| 18 |
Hb_000521_360 |
0.1379037325 |
- |
- |
PREDICTED: beta-glucosidase 17-like [Jatropha curcas] |
| 19 |
Hb_000649_280 |
0.1382764527 |
- |
- |
PREDICTED: F-box protein PP2-B10-like [Pyrus x bretschneideri] |
| 20 |
Hb_000108_090 |
0.1397172236 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB5-like [Jatropha curcas] |