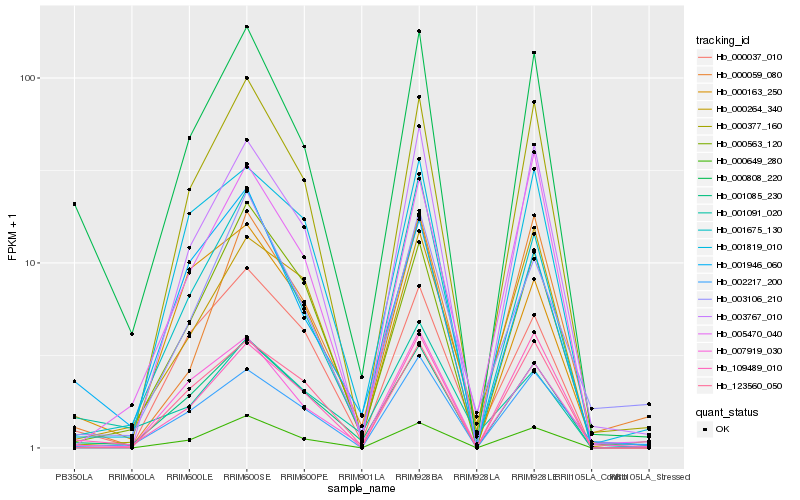
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000808_220 |
0.0 |
- |
- |
PREDICTED: pyrophosphate-energized vacuolar membrane proton pump 1-like [Jatropha curcas] |
| 2 |
Hb_001085_230 |
0.1142768352 |
- |
- |
PREDICTED: receptor like protein kinase S.2 [Jatropha curcas] |
| 3 |
Hb_109489_010 |
0.118895674 |
- |
- |
multidrug resistance-associated protein 2, 6 (mrp2, 6), abc-transoprter, putative [Ricinus communis] |
| 4 |
Hb_001946_060 |
0.1274681325 |
- |
- |
PREDICTED: TBC1 domain family member 13-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_001675_130 |
0.1359707002 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein GLABRA 2 [Jatropha curcas] |
| 6 |
Hb_007919_030 |
0.1411647729 |
- |
- |
- |
| 7 |
Hb_000377_160 |
0.1447081715 |
- |
- |
PREDICTED: oligopeptide transporter 3 [Jatropha curcas] |
| 8 |
Hb_000649_280 |
0.1510495492 |
- |
- |
PREDICTED: F-box protein PP2-B10-like [Pyrus x bretschneideri] |
| 9 |
Hb_003767_010 |
0.1522135423 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD11 [Jatropha curcas] |
| 10 |
Hb_002217_200 |
0.1540933541 |
- |
- |
PREDICTED: gibberellin 2-beta-dioxygenase 2 [Jatropha curcas] |
| 11 |
Hb_000037_010 |
0.1554956253 |
- |
- |
PREDICTED: cation/H(+) antiporter 18-like [Jatropha curcas] |
| 12 |
Hb_000163_250 |
0.1599961342 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 13 |
Hb_123560_050 |
0.160098612 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g24010 [Jatropha curcas] |
| 14 |
Hb_000563_120 |
0.1606008024 |
- |
- |
hypothetical protein JCGZ_18936 [Jatropha curcas] |
| 15 |
Hb_005470_040 |
0.1678177107 |
- |
- |
hypothetical protein POPTR_0015s09760g [Populus trichocarpa] |
| 16 |
Hb_000059_080 |
0.1685990111 |
- |
- |
PREDICTED: uncharacterized protein LOC105629177 [Jatropha curcas] |
| 17 |
Hb_001091_020 |
0.1710296236 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 18 |
Hb_001819_010 |
0.17146972 |
- |
- |
PREDICTED: potassium transporter 8 [Jatropha curcas] |
| 19 |
Hb_000264_340 |
0.1725540562 |
- |
- |
PREDICTED: callose synthase 11-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_003106_210 |
0.1786813415 |
- |
- |
PREDICTED: UDP-glycosyltransferase 74B1-like [Jatropha curcas] |