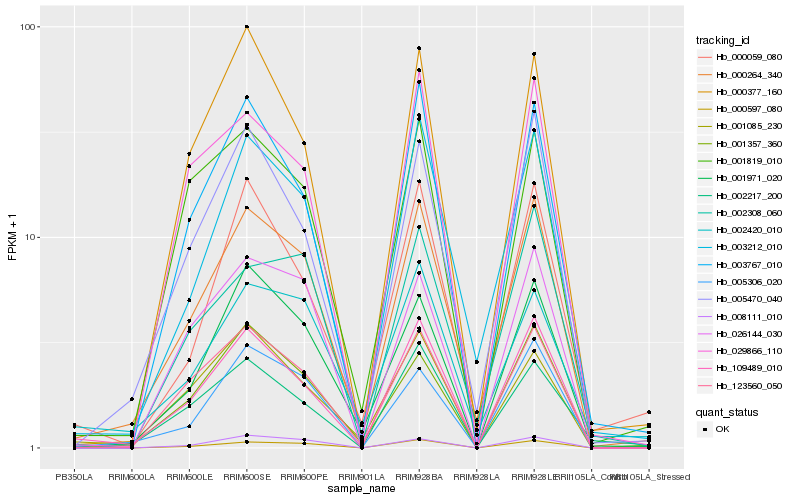
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000264_340 |
0.0 |
- |
- |
PREDICTED: callose synthase 11-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_003212_010 |
0.0811460845 |
- |
- |
sucrose transporter 4 [Hevea brasiliensis] |
| 3 |
Hb_005470_040 |
0.098091289 |
- |
- |
hypothetical protein POPTR_0015s09760g [Populus trichocarpa] |
| 4 |
Hb_109489_010 |
0.1076995283 |
- |
- |
multidrug resistance-associated protein 2, 6 (mrp2, 6), abc-transoprter, putative [Ricinus communis] |
| 5 |
Hb_123560_050 |
0.1078858544 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g24010 [Jatropha curcas] |
| 6 |
Hb_000597_080 |
0.1107491979 |
- |
- |
PREDICTED: 7-deoxyloganetic acid glucosyltransferase-like isoform X2 [Jatropha curcas] |
| 7 |
Hb_001971_020 |
0.1125639933 |
- |
- |
PREDICTED: alpha-dioxygenase 2 [Populus euphratica] |
| 8 |
Hb_001357_360 |
0.1140777042 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 9 |
Hb_005306_020 |
0.1205010313 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB86 [Jatropha curcas] |
| 10 |
Hb_003767_010 |
0.1242362352 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD11 [Jatropha curcas] |
| 11 |
Hb_002420_010 |
0.1252180491 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 12 |
Hb_000377_160 |
0.1263539235 |
- |
- |
PREDICTED: oligopeptide transporter 3 [Jatropha curcas] |
| 13 |
Hb_002308_060 |
0.1277302806 |
- |
- |
PREDICTED: cytochrome P450 704C1-like [Jatropha curcas] |
| 14 |
Hb_008111_010 |
0.1288591956 |
- |
- |
hypothetical protein VITISV_021527 [Vitis vinifera] |
| 15 |
Hb_000059_080 |
0.1291116499 |
- |
- |
PREDICTED: uncharacterized protein LOC105629177 [Jatropha curcas] |
| 16 |
Hb_001819_010 |
0.1309837658 |
- |
- |
PREDICTED: potassium transporter 8 [Jatropha curcas] |
| 17 |
Hb_029866_110 |
0.1346873239 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: ABC transporter C family member 10-like [Jatropha curcas] |
| 18 |
Hb_001085_230 |
0.1351846647 |
- |
- |
PREDICTED: receptor like protein kinase S.2 [Jatropha curcas] |
| 19 |
Hb_026144_030 |
0.1352188448 |
- |
- |
PREDICTED: high affinity nitrate transporter 2.5 [Jatropha curcas] |
| 20 |
Hb_002217_200 |
0.1372702637 |
- |
- |
PREDICTED: gibberellin 2-beta-dioxygenase 2 [Jatropha curcas] |