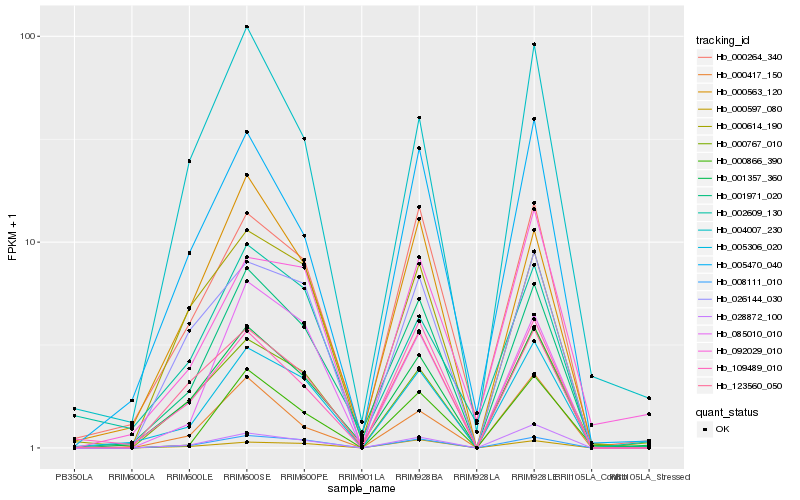
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005306_020 |
0.0 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB86 [Jatropha curcas] |
| 2 |
Hb_008111_010 |
0.1003489921 |
- |
- |
hypothetical protein VITISV_021527 [Vitis vinifera] |
| 3 |
Hb_001971_020 |
0.1064238086 |
- |
- |
PREDICTED: alpha-dioxygenase 2 [Populus euphratica] |
| 4 |
Hb_001357_360 |
0.1109670545 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 5 |
Hb_028872_100 |
0.1171848423 |
- |
- |
PREDICTED: uncharacterized protein LOC102707106 [Oryza brachyantha] |
| 6 |
Hb_000264_340 |
0.1205010313 |
- |
- |
PREDICTED: callose synthase 11-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_000767_010 |
0.1250271297 |
- |
- |
PREDICTED: geraniol 8-hydroxylase-like isoform X2 [Solanum lycopersicum] |
| 8 |
Hb_085010_010 |
0.1327026094 |
- |
- |
PREDICTED: MLO-like protein 3 isoform X2 [Vitis vinifera] |
| 9 |
Hb_026144_030 |
0.1329053001 |
- |
- |
PREDICTED: high affinity nitrate transporter 2.5 [Jatropha curcas] |
| 10 |
Hb_005470_040 |
0.1345515532 |
- |
- |
hypothetical protein POPTR_0015s09760g [Populus trichocarpa] |
| 11 |
Hb_109489_010 |
0.1418374398 |
- |
- |
multidrug resistance-associated protein 2, 6 (mrp2, 6), abc-transoprter, putative [Ricinus communis] |
| 12 |
Hb_000597_080 |
0.1467626219 |
- |
- |
PREDICTED: 7-deoxyloganetic acid glucosyltransferase-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_000417_150 |
0.1474835665 |
- |
- |
hypothetical protein JCGZ_21586 [Jatropha curcas] |
| 14 |
Hb_000866_390 |
0.1491440431 |
- |
- |
PREDICTED: O-glucosyltransferase rumi homolog [Jatropha curcas] |
| 15 |
Hb_092029_010 |
0.1520544527 |
- |
- |
Disease resistance protein RPM1, putative [Ricinus communis] |
| 16 |
Hb_123560_050 |
0.1522074757 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g24010 [Jatropha curcas] |
| 17 |
Hb_000563_120 |
0.153680583 |
- |
- |
hypothetical protein JCGZ_18936 [Jatropha curcas] |
| 18 |
Hb_000614_190 |
0.1568529479 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: putative ABC transporter C family member 15 [Jatropha curcas] |
| 19 |
Hb_002609_130 |
0.1572826752 |
- |
- |
PREDICTED: cytochrome P450 86A1 [Jatropha curcas] |
| 20 |
Hb_004007_230 |
0.1581704951 |
- |
- |
PREDICTED: putative formamidase C869.04 isoform X1 [Jatropha curcas] |