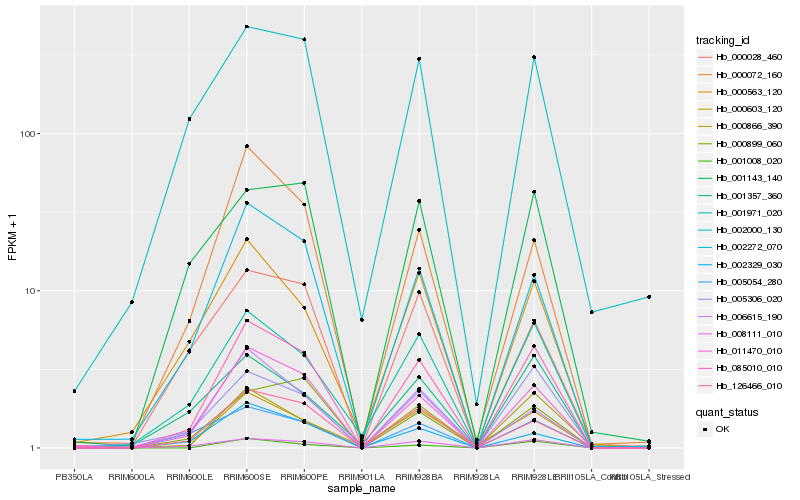
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_085010_010 |
0.0 |
- |
- |
PREDICTED: MLO-like protein 3 isoform X2 [Vitis vinifera] |
| 2 |
Hb_011470_010 |
0.0848065659 |
- |
- |
hypothetical protein CISIN_1g022501mg [Citrus sinensis] |
| 3 |
Hb_008111_010 |
0.0987517596 |
- |
- |
hypothetical protein VITISV_021527 [Vitis vinifera] |
| 4 |
Hb_126466_010 |
0.1157072727 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 5 |
Hb_002272_070 |
0.1228417247 |
- |
- |
PREDICTED: keratin, type I cytoskeletal 9 [Jatropha curcas] |
| 6 |
Hb_001971_020 |
0.1314835113 |
- |
- |
PREDICTED: alpha-dioxygenase 2 [Populus euphratica] |
| 7 |
Hb_005306_020 |
0.1327026094 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB86 [Jatropha curcas] |
| 8 |
Hb_005054_280 |
0.1405191613 |
transcription factor |
TF Family: HB |
Protein WUSCHEL, putative [Ricinus communis] |
| 9 |
Hb_000866_390 |
0.1412532895 |
- |
- |
PREDICTED: O-glucosyltransferase rumi homolog [Jatropha curcas] |
| 10 |
Hb_001008_020 |
0.1456159871 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 11 |
Hb_000603_120 |
0.1492481871 |
- |
- |
Protein kinase APK1A, chloroplast precursor, putative [Ricinus communis] |
| 12 |
Hb_000072_160 |
0.1540448781 |
- |
- |
proline oxidase [Manihot esculenta] |
| 13 |
Hb_000563_120 |
0.1576897536 |
- |
- |
hypothetical protein JCGZ_18936 [Jatropha curcas] |
| 14 |
Hb_006615_190 |
0.1596651899 |
- |
- |
UDP-glycosyltransferase 1 [Linum usitatissimum] |
| 15 |
Hb_002000_130 |
0.1637049143 |
- |
- |
PREDICTED: abscisic stress-ripening protein 2-like [Eucalyptus grandis] |
| 16 |
Hb_001143_140 |
0.1648682085 |
- |
- |
Aldehyde dehydrogenase, putative [Ricinus communis] |
| 17 |
Hb_000899_060 |
0.1664682062 |
- |
- |
PREDICTED: uncharacterized protein LOC105793631 [Gossypium raimondii] |
| 18 |
Hb_002329_030 |
0.1672422682 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 2-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_000028_460 |
0.1692811217 |
- |
- |
hypothetical protein POPTR_0015s05340g [Populus trichocarpa] |
| 20 |
Hb_001357_360 |
0.1701761786 |
- |
- |
zinc finger protein, putative [Ricinus communis] |