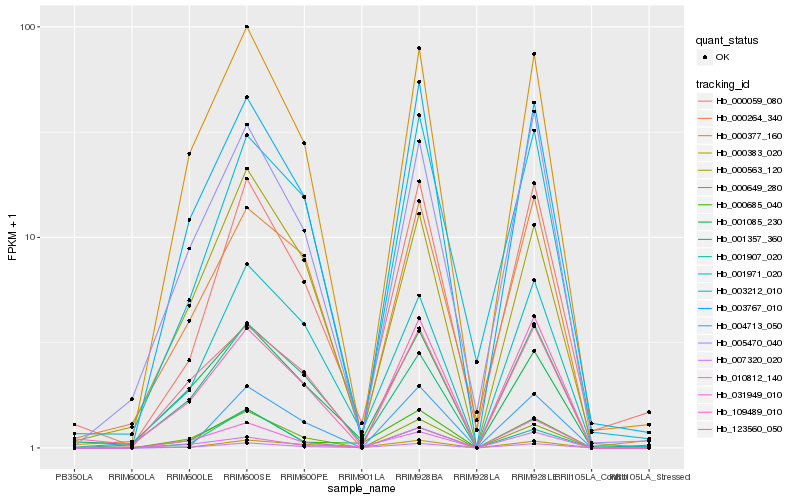
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007320_020 |
0.0 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 isoform X2 [Populus euphratica] |
| 2 |
Hb_000685_040 |
0.0942362612 |
- |
- |
hypothetical protein OsJ_17676 [Oryza sativa Japonica Group] |
| 3 |
Hb_001971_020 |
0.1181492989 |
- |
- |
PREDICTED: alpha-dioxygenase 2 [Populus euphratica] |
| 4 |
Hb_000059_080 |
0.1207933322 |
- |
- |
PREDICTED: uncharacterized protein LOC105629177 [Jatropha curcas] |
| 5 |
Hb_003767_010 |
0.130495027 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD11 [Jatropha curcas] |
| 6 |
Hb_010812_140 |
0.1344199657 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 7 |
Hb_000377_160 |
0.1349424354 |
- |
- |
PREDICTED: oligopeptide transporter 3 [Jatropha curcas] |
| 8 |
Hb_000649_280 |
0.1392457852 |
- |
- |
PREDICTED: F-box protein PP2-B10-like [Pyrus x bretschneideri] |
| 9 |
Hb_000383_020 |
0.1463528882 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 10 |
Hb_005470_040 |
0.1475711685 |
- |
- |
hypothetical protein POPTR_0015s09760g [Populus trichocarpa] |
| 11 |
Hb_001357_360 |
0.1506635782 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 12 |
Hb_031949_010 |
0.1514026649 |
- |
- |
- |
| 13 |
Hb_004713_050 |
0.1538659419 |
- |
- |
RecName: Full=Beta-amyrin synthase [Betula platyphylla] |
| 14 |
Hb_109489_010 |
0.1540130387 |
- |
- |
multidrug resistance-associated protein 2, 6 (mrp2, 6), abc-transoprter, putative [Ricinus communis] |
| 15 |
Hb_003212_010 |
0.1547934371 |
- |
- |
sucrose transporter 4 [Hevea brasiliensis] |
| 16 |
Hb_001085_230 |
0.1550958251 |
- |
- |
PREDICTED: receptor like protein kinase S.2 [Jatropha curcas] |
| 17 |
Hb_123560_050 |
0.1584989174 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g24010 [Jatropha curcas] |
| 18 |
Hb_000264_340 |
0.1596225019 |
- |
- |
PREDICTED: callose synthase 11-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_000563_120 |
0.1597911768 |
- |
- |
hypothetical protein JCGZ_18936 [Jatropha curcas] |
| 20 |
Hb_001907_020 |
0.1682767002 |
- |
- |
PREDICTED: receptor-like protein 12 [Vitis vinifera] |