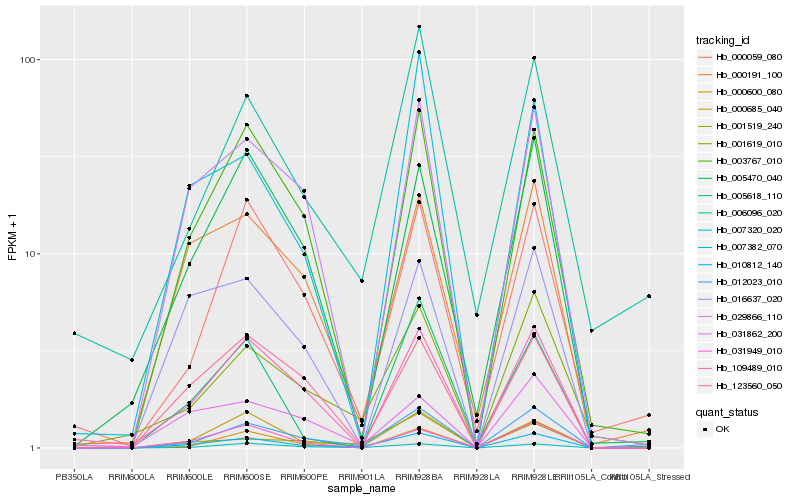
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010812_140 |
0.0 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 2 |
Hb_000685_040 |
0.1201587248 |
- |
- |
hypothetical protein OsJ_17676 [Oryza sativa Japonica Group] |
| 3 |
Hb_007320_020 |
0.1344199657 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 isoform X2 [Populus euphratica] |
| 4 |
Hb_012023_010 |
0.1406811224 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001619_010 |
0.1460297974 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET2a-like [Jatropha curcas] |
| 6 |
Hb_003767_010 |
0.1504256325 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD11 [Jatropha curcas] |
| 7 |
Hb_031949_010 |
0.1561383039 |
- |
- |
- |
| 8 |
Hb_029866_110 |
0.1594655021 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: ABC transporter C family member 10-like [Jatropha curcas] |
| 9 |
Hb_000600_080 |
0.1645926574 |
- |
- |
- |
| 10 |
Hb_109489_010 |
0.1656143569 |
- |
- |
multidrug resistance-associated protein 2, 6 (mrp2, 6), abc-transoprter, putative [Ricinus communis] |
| 11 |
Hb_000191_100 |
0.1684888799 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 12 |
Hb_005470_040 |
0.1699009486 |
- |
- |
hypothetical protein POPTR_0015s09760g [Populus trichocarpa] |
| 13 |
Hb_001519_240 |
0.1738630367 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 14 |
Hb_016637_020 |
0.1760376377 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB5-like [Jatropha curcas] |
| 15 |
Hb_007382_070 |
0.1768793481 |
- |
- |
PREDICTED: MAPK-interacting and spindle-stabilizing protein-like [Jatropha curcas] |
| 16 |
Hb_005618_110 |
0.1773801896 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 17 |
Hb_031862_200 |
0.1794998546 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At3g14460 [Vitis vinifera] |
| 18 |
Hb_006096_020 |
0.1797184289 |
- |
- |
Os06g0343500 [Oryza sativa Japonica Group] |
| 19 |
Hb_123560_050 |
0.1802652141 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g24010 [Jatropha curcas] |
| 20 |
Hb_000059_080 |
0.1811406077 |
- |
- |
PREDICTED: uncharacterized protein LOC105629177 [Jatropha curcas] |