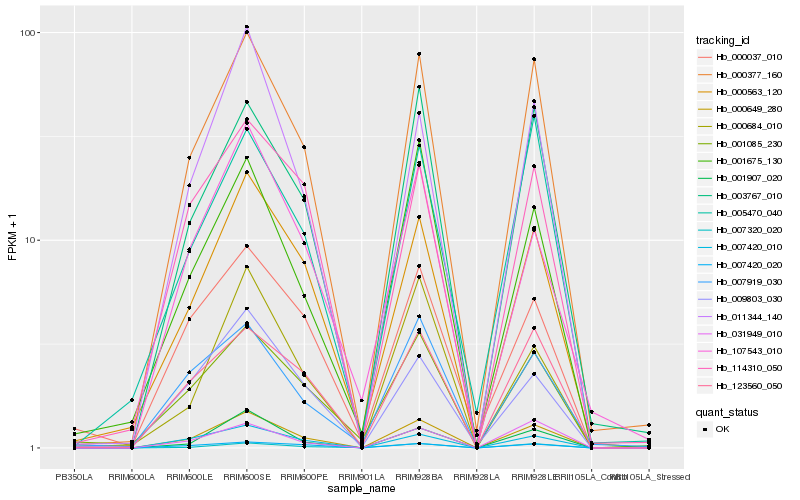
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000649_280 |
0.0 |
- |
- |
PREDICTED: F-box protein PP2-B10-like [Pyrus x bretschneideri] |
| 2 |
Hb_000377_160 |
0.0546370388 |
- |
- |
PREDICTED: oligopeptide transporter 3 [Jatropha curcas] |
| 3 |
Hb_003767_010 |
0.0981388544 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD11 [Jatropha curcas] |
| 4 |
Hb_000563_120 |
0.0994716081 |
- |
- |
hypothetical protein JCGZ_18936 [Jatropha curcas] |
| 5 |
Hb_007420_010 |
0.1060244018 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X2 [Jatropha curcas] |
| 6 |
Hb_001907_020 |
0.1121395373 |
- |
- |
PREDICTED: receptor-like protein 12 [Vitis vinifera] |
| 7 |
Hb_123560_050 |
0.1124490116 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g24010 [Jatropha curcas] |
| 8 |
Hb_011344_140 |
0.114158428 |
- |
- |
PREDICTED: geraniol 8-hydroxylase-like [Jatropha curcas] |
| 9 |
Hb_001085_230 |
0.1221450407 |
- |
- |
PREDICTED: receptor like protein kinase S.2 [Jatropha curcas] |
| 10 |
Hb_007420_020 |
0.1240474672 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 11 |
Hb_031949_010 |
0.1277210692 |
- |
- |
- |
| 12 |
Hb_001675_130 |
0.1285987024 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein GLABRA 2 [Jatropha curcas] |
| 13 |
Hb_009803_030 |
0.1345880978 |
- |
- |
PREDICTED: probable receptor protein kinase TMK1 [Jatropha curcas] |
| 14 |
Hb_114310_050 |
0.1360546015 |
- |
- |
Iron(III)-zinc(II) purple acid phosphatase precursor family protein [Populus trichocarpa] |
| 15 |
Hb_000684_010 |
0.1380566961 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 16 |
Hb_007919_030 |
0.1382764527 |
- |
- |
- |
| 17 |
Hb_007320_020 |
0.1392457852 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 isoform X2 [Populus euphratica] |
| 18 |
Hb_005470_040 |
0.1398808338 |
- |
- |
hypothetical protein POPTR_0015s09760g [Populus trichocarpa] |
| 19 |
Hb_107543_010 |
0.1399898559 |
- |
- |
Cysteine-rich RLK 10, putative [Theobroma cacao] |
| 20 |
Hb_000037_010 |
0.140192269 |
- |
- |
PREDICTED: cation/H(+) antiporter 18-like [Jatropha curcas] |